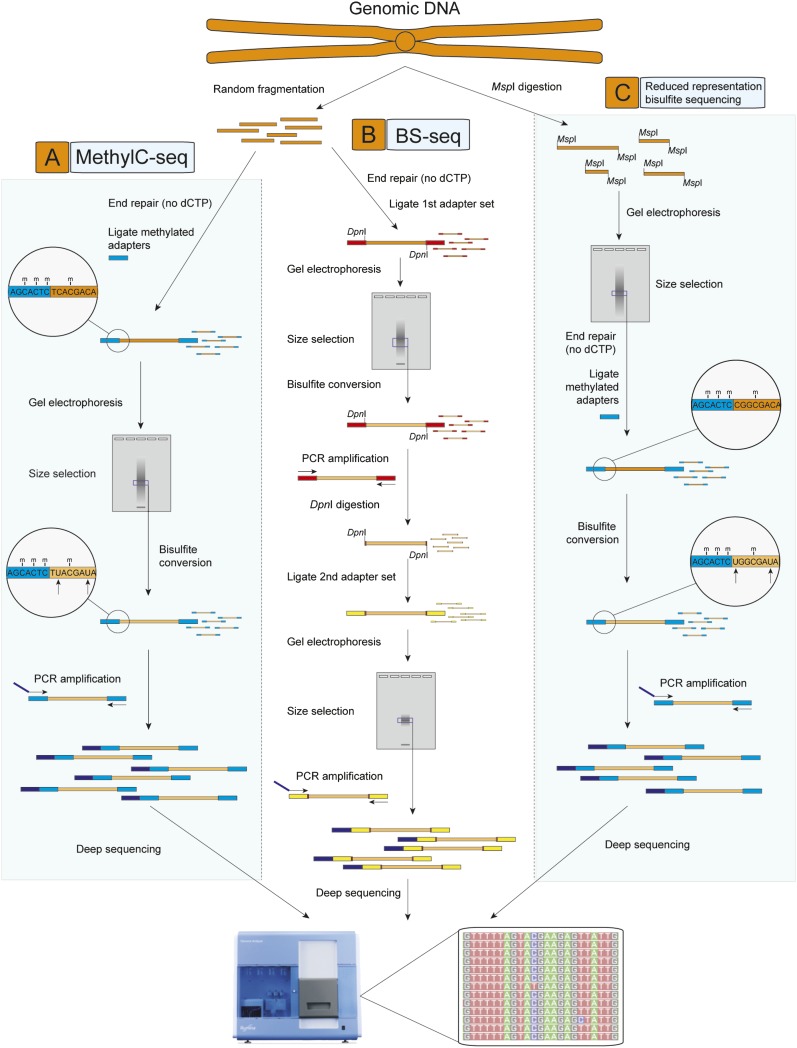
Figure 1.
Techniques for genome-wide sequencing of cytosine methylation sites. Three techniques used recently to generate bisulfite (BS) sequencing libraries compatible with next-generation sequencing are depicted. (A) MethylC-seq (Lister et al. 2008). Double-stranded universal adapter sequences in which all cytosines are methylated are ligated to fragmented genomic DNA. Sodium bisulfite treatment converts unmethylated cytosines to thymine, after which library yield enrichment by PCR with primers complementary to the universal adapter sequences produces the final library that can be sequenced. (B) BS-seq (Cokus et al. 2008). Ligation of a first set of double-stranded adaptors that contained methylated adenine bases within DpnI restriction sites close to the site of ligation with genomic DNA. After BS conversion, PCR is performed using primers complementary to the converted adapter sequences, yielding double-stranded DNA that is digested with DpnI to remove only the first adapter set. Sequencing adapters are subsequently ligated to the double-stranded BS-converted genomic DNA fragments, and PCR with primers complementary to the adapters performed to yield a sequencing library. (C) Reduced representation BS sequencing (RRBS) (Meissner et al. 2008). Genomic DNA is first digested by the methylation-insensitive MspI restriction enzyme, which cleaves the phosphodiester bond upstream of the CpG dinuclotide in its CCGG recognition element. Digested DNA is then separated by gel electrophoresis, and one or more specific size fractions are selected. The size-selected DNA is then end repaired, ligated to double-stranded methylated sequencing adapters (as described above for MethylC-seq), BS converted, and amplified by PCR with primers complementary to the adapter sequences.