Figure 5.
SERF1a Binds to the C-Terminal Region of aSyn
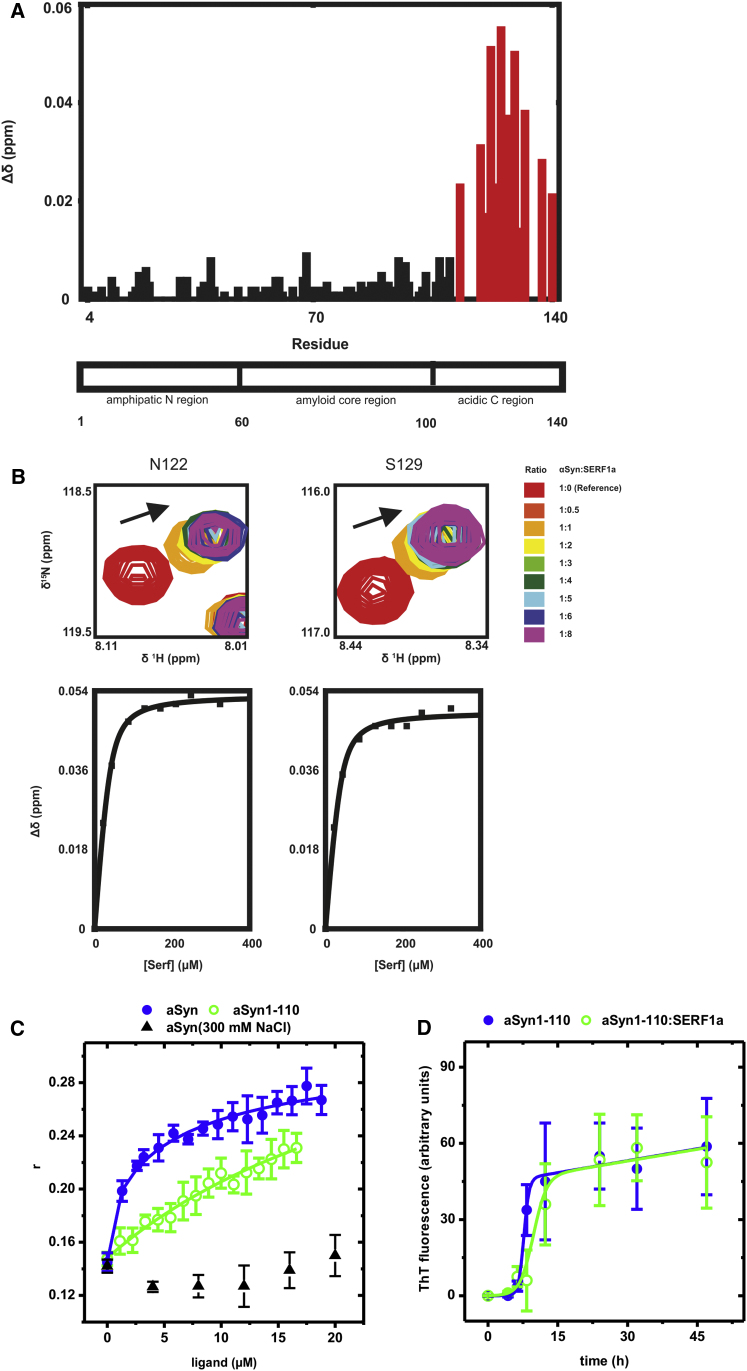
(A) aSyn 1H,15N-HSQC-NMR perturbation diagram showing chemical shift changes Δδ after the addition of SERF1a. The largest changes were localized to the C-terminal region, between the amino acids Gly111-Ala140 (red columns).
(B) Upper: Detail view of well-resolved and large chemical shift changes that occur for amino acid signals Asp122 and Ser129, upon titration with SERF1a (arrows). Lower panels: The corresponding binding curves yielded dissociation constants KD = 7.59 ± 1.87 μM (Asp122) and 9.81 ± 1.06 μM (Ser129).
(C) Fluorescence anisotropy binding curves of Atto550-SERF1a upon titration with aSyn (closed blue circle; KD = 8.04 ± 2.09 μM) and aSyn1-110 (open green circle; nonsaturable binding, extrapolated KD > 1 mM). The interaction between Atto550-SERF1a with aSyn was abolished by increasing the ionic strength of the solution to 300 mM NaCl (closed upward black triangle), pointing to a predominantly hydrophilic binding mode.
(D) Amyloid fiber growth of the truncation mutant aSyn1-110 was insensitive to SERF1a: amyloid kinetics of aSyn1-110 in the absence (closed blue circle) and in the presence (open green circle) of SERF1a. Note that this truncation intrinsically improves the amyloid properties of aSyn (tm = 7.70 ± 0.18 hr; tl = 6.56 ± 0.83 hr; kapp = 1.75 ± 0.17 hr−; see also Hoyer et al., 2004 and Levitan et al., 2011).
See also Figure S4. The error bars are mean averages, including SEM of three independent experiments.