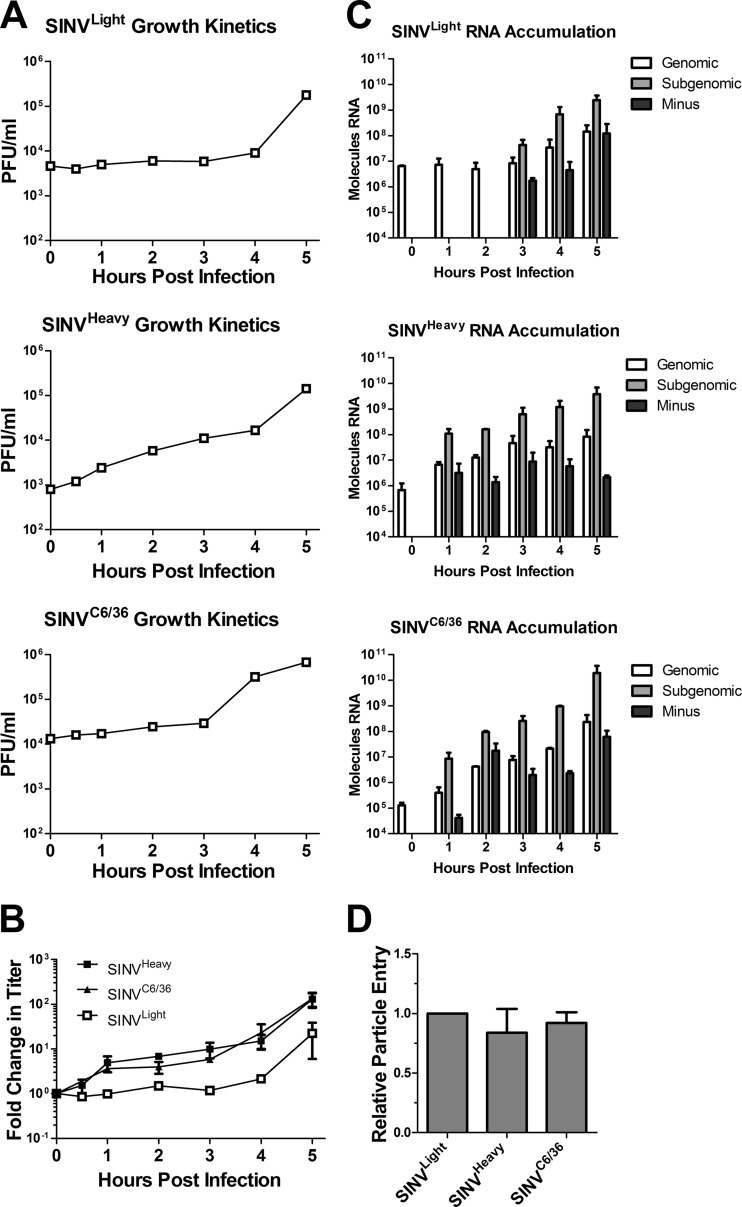
Fig 3.
Viral growth kinetics and RNA synthesis and accumulation are enhanced during infection with SINVHeavy or SINVC6/36. (A) Monolayers of BHK-21 cells were infected with the indicated SINV subpopulations at an MOI of 10 PFU/cell as described in Materials and Methods. At the indicated times postinfection, cell supernatants were collected, and titers were determined. (B) The average fold change in titer observed for each of the individual SINV subpopulations during infection of BHK-21 cells (from panel A) is plotted against time. Data are mean values for at least two independent biological replicates. Error bars represent standard deviations. (C) Monolayers of BHK-21 cells were infected with the indicated SINV subpopulations at an MOI of 10 PFU/cell as described in Materials and Methods. At the indicated time points postinfection, total cellular RNA was extracted and was used to determine the amount of viral RNA by qRT-PCR. The absolute quantities of the viral RNA species were calculated using standard curves. The absence of a value means that the signal was either below the threshold of detection or below the background of the sample. Data shown are representative of three independent biological replicates. (D) BHK-21 cells were infected with equal numbers of SINVLight, SINVHeavy, or SINVC6/36 particles (in contrast to equal numbers of infectious particles, as for panels A to C). After extensive washing, the viral RNA content was quantified as for panel A. Data are mean values for two independent biological replicates. Error bars represent standard deviations.