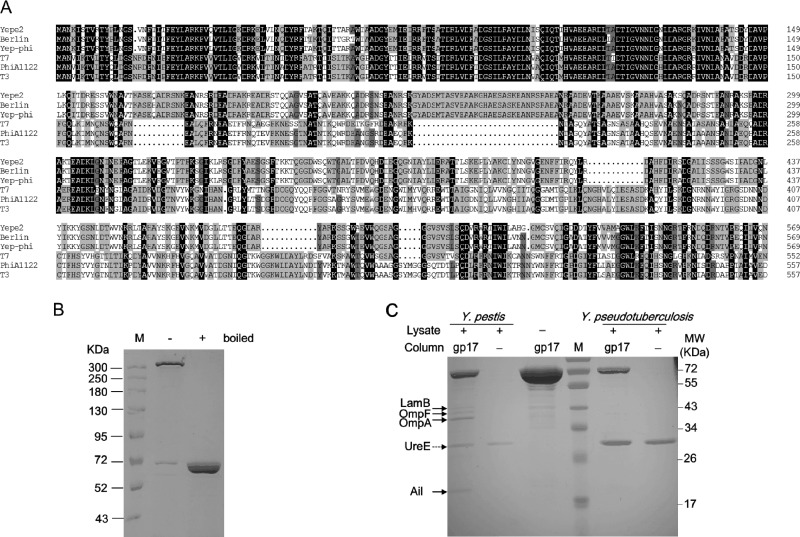
Fig 2.
(A) Alignment of tail fiber proteins. Identical residues are shaded in black, residues sharing >75% homology are shaded in deep gray, and those sharing >50% homology are shaded in light gray. Gaps (indicated by dotted lines) were introduced into the sequences to maximize the alignments. Numbering is based on the N-terminal methionine. Multiple sequence alignment was analyzed and edited with DNAman software (Lynnonon Biosoft, Quebec, Canada). The names of phages are indicated on the left. (B) SDS-PAGE gel showing the SDS-resistant multimer formed by gp17. Symbols “+” and “−” above the gel denote the purified protein boiled previously and unboiled, respectively. (C) Identification of cell wall-anchored protein targets of gp17 by affinity chromatography. The protein bands found to interact specifically with gp17 are indicated by black arrows. UreE protein binding to the nickel column is denoted by a dotted arrow.