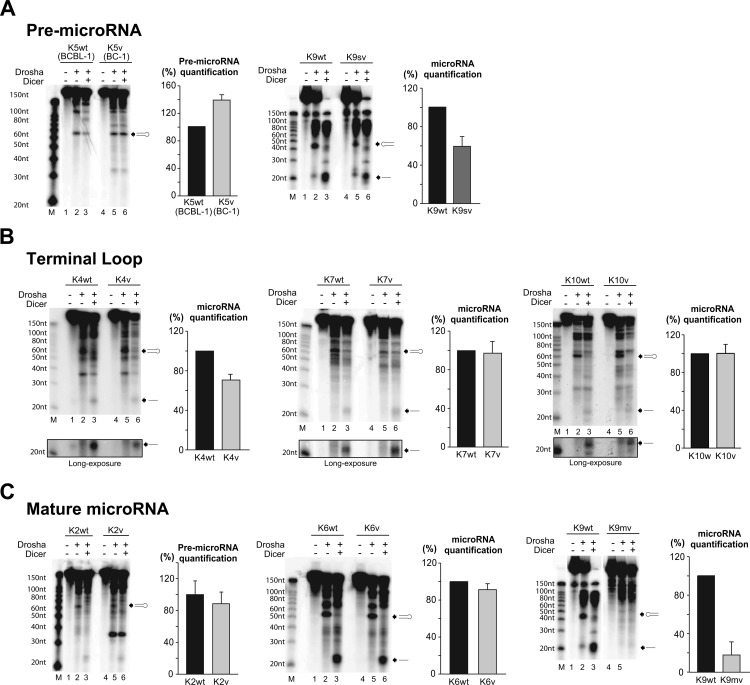
Fig 4.
A. In vitro maturation assay for miR-K12-5 and -9. These microRNA variants have SNPs in pre-microRNA. (First and third panels) Products on urea-polyacrylamide gel. Black arrows, pre-microRNA and microRNA produced from each processing; lanes M, molecular size markers. (Second and fourth panels) Graphs of pre-microRNA quantification normalized by the levels for the wt. Results are shown as percentages ± standard deviations from three independent experiments. (B) In vitro maturation assay for miR-K12-4, miR-K12-7, and miR-K12-10. The variants of these microRNAs have SNPs in different positions in the terminal loop of pre-microRNA. (First, third, and fifth panels) Products on urea-polyacrylamide gels. Black arrows, pre-microRNA and microRNA produced from each processing; lanes M, molecular size markers. (Second, fourth, and sixth panels) Graphs of pre-microRNA quantification normalized by the levels for the wt. Results are shown as percentages ± standard deviations from three independent experiments. (C) In vitro maturation assay for miR-K12-2, miR-K12-6, and miR-K12-9mv. The variants of these microRNAs have an SNP or SNPs within mature microRNA sequences. (First, third, and fifth panels) Products on urea-polyacrylamide gels. Black arrows, pre-microRNA produced; lanes M, molecular size markers. (Second, fourth, and sixth panels) Graphs of pre-microRNA quantification normalized by the levels for the wt. Results are shown as percentages ± standard deviations from three independent experiments.