Fig 1.
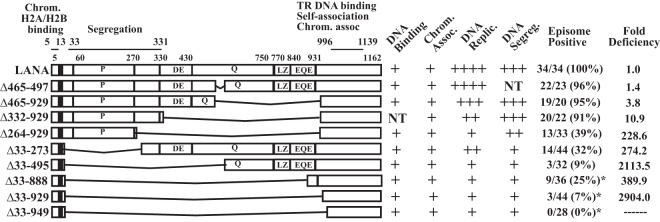
Schematic diagram of KSHV LANA and LANA deletion mutants. Indicated are the proline-rich region (P), the aspartate and glutamate (DE), glutamine (Q), and glutamate and glutamine (EQE) regions, and the putative leucine zipper (LZ). The DE, Q, EQE, and LZ regions all contain repeat elements. The shaded area represents region I of the N-terminal nuclear localization signal (NLS) within amino acids 24 to 30 (20, 69). The C-terminal portion of LANA can also localize to nuclei, but an NLS has not been precisely mapped. Amino acids 5 to 13 mediate chromosome association through interaction with histones H2A/H2B. Amino acids 996 to 1139 contain TR DNA binding, self-association, and chromosome association functions. The capabilities for TR DNA binding, mitotic chromosome association, DNA replication, DNA segregation (as suggested by retention of p2TR-ΔRE-GFP), and episome persistence for each of the constructs are shown on the right. For episome persistence, fractions indicate the number of G418-resistant cell lines with episomes divided by the total number of G418-resistant cell lines assayed by Gardella analysis; percentages are given in parentheses. Fold deficiencies in episome maintenance ability were determined by dividing the value for each mutant in Table 1 by that for WT LANA. NT, not tested. LANA deletion mutants marked with asterisks, and their abilities to bind TR DNA, associate with chromosomes, and maintain p8TR episomes, have been described previously (28), but their TR DNA replication and segregation capabilities were further investigated here.
