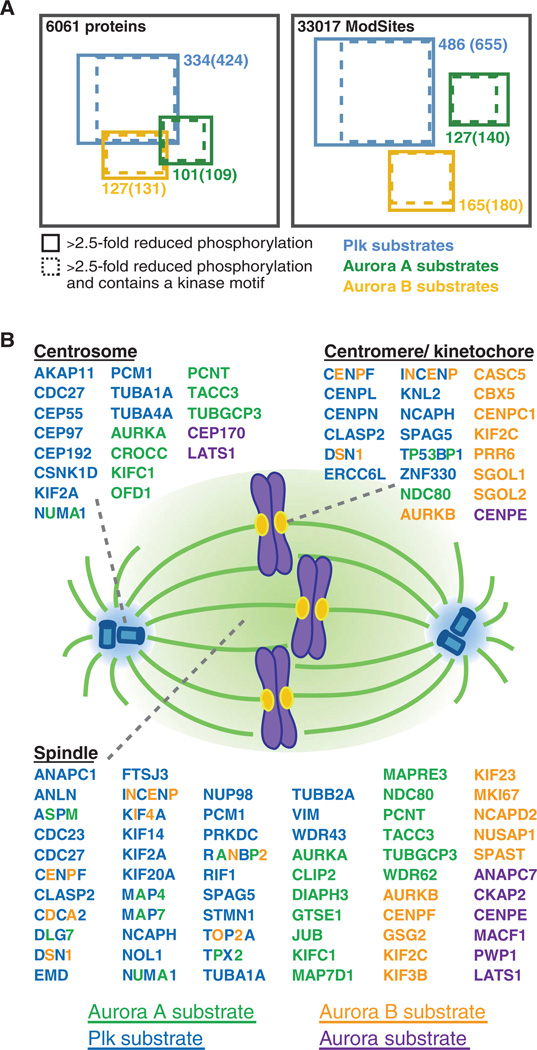
Fig. 5.
Candidate Aurora A, Aurora B, and Plk substrates and their spindle localization. (A) Diagram of proteins and unique ModSites identified in HeLa cells by quantitative chemical phosphoproteomics. Total number of identifications is indicated in black. Blue squares indicate candidate Plk substrates; green squares, candidate Aurora A substrates; and orange squares, candidate Aurora B substrates. Number of proteins and ModSites in each group that displayed >2.5-fold reduction in phosphorylation after inhibitor treatment, as well as those that contained a minimal kinase motif (in parentheses) is given. (B) Depiction of candidate Aurora A (green), Aurora B (orange), Aurora ambiguous (purple), and Plk (blue) substrates based on their subcellular localization to centromeres or kinetochores, centrosomes, and the spindle. Proteins with more than one colored letter are substrates of more than one of these kinases. The number of colored letters is not an indication of the number of phosphorylation sites matched to a particular kinase.