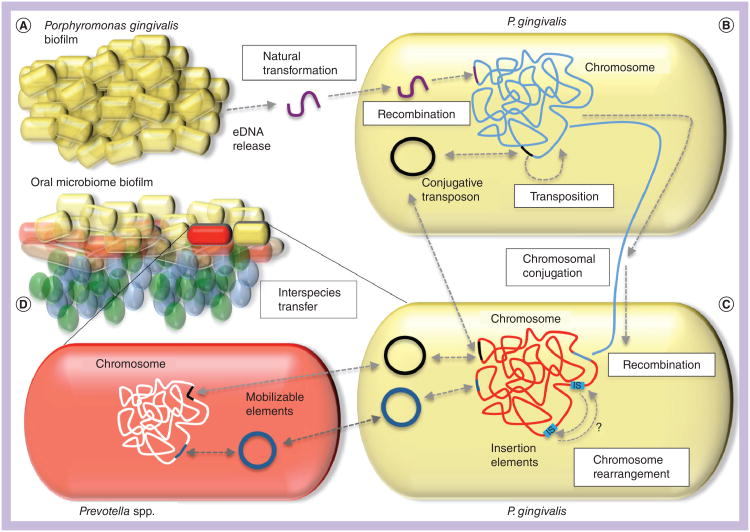
Figure 4. Mechanisms of DNA transfer in Porphyromonas gingivalis.
Porphyromonas gingivalis resides in dental plaque biofilms in the presence of diverse bacterial species and multiple strains of P. gingivalis may coexist within the same biofilm community. (A)P. gingivalis biofilms have been shown to release eDNA (purple) into the biofilm matrix. This DNA can be taken up by other strains of P. gingivalis by natural transformation. This process requires the comF gene in the recipient strain; comF proteins provide the molecular motor to bring DNA across the bacterial membrane. DNA that is homologous to the host cell genome may be integrated into the chromosome by homologous recombination. (B) Some strains of P. gingivalis contain conjugative transposons that are normally integrated into the chromosome (black line), but may excise and form a circular intermediate prior to conjugative transfer to a new host cell (black circle). Conjugative transposon cTnPg1 has been shown to transfer between P. gingivalis strains, is able to transfer into other closely related species such as Prevotella (D), and can also mobilize plasmids and other genetic elements between species (dark blue circles). (C) Conjugation can also mediate the transfer of chromosomal DNA between strains of P. gingivalis. This process is dependent on conjugative transposons or other traAQ-containing elements present in the donor strain and homologous recombination in the recipient. Allelic replacement of recipient DNA (red) with donor DNA (blue line) is mediated by homologous recombination. Finally, insertion elements (light blue boxes) are present in P. gingivalis and are capable of transposing into genes or their promoter regions, thus influencing expression. They are also likely responsible for the large scale genomic rearrangements that can be detected by comparison of multiple genome sequences. (D) cTnPg1 can be transferred to the closely related genus Prevotella. eDNA: Extracellular DNA.