Abstract
Ethylene is an important plant hormone that regulates developmental processes in plants. The ethylene biosynthesis pathway is a highly regulated process at both the transcriptional and post-translational level. The transcriptional regulation of these ethylene biosynthesis genes is well known. However, post-translational modifications of the key ethylene biosynthesis enzyme 1-aminocyclopropane-1-carboxylate (ACC) synthase (ACS) are little understood. In vitro kinase assays were conducted on the type III ACS, AtACS7, fusion protein and peptides to determine whether the AtACS7 protein can be phosphorylated by calcium-dependent protein kinase (CDPK). AtACS7 was phosphorylated at Ser216, Thr296, and Ser299 by AtCDPK16 in vitro. To investigate further the function of the ACS7 gene in Arabidopsis, an acs7-1 loss-of-function mutant was isolated. The acs7-1 mutant exhibited less sensitivity to the inhibition of root gravitropism by treatment with the calcium chelator ethylene glycol tetraacetic acid (EGTA). Seedlings were treated with gradient concentrations of ACC. The results showed that a certain concentration of ethylene enhanced the gravity response. Moreover, the acs7-1 mutant was less sensitive to inhibition of the gravity response by treatment with the auxin polar transport inhibitor 1-naphthylphthalamic acid, but exogenous ACC application recovered root gravitropism. Altogether, the results indicate that AtACS7 is involved in root gravitropism in a calcium-dependent manner in Arabidopsis.
Key words: 14-3-3, ACS, calcium, CDPK, ethylene, phosphorylation, root gravitropism.
Introduction
Gravity is one of the most important environmental cues that control growth direction (Morita, 2010). Shoots generally grow upward (i.e. negative gravitropism) and roots grow downward (i.e. positive gravitropism). It is unknown how plants receive and respond to the gravity signal. One widely accepted theory is that starch-accumulating amyloplast movement along the gravity vector within gravity-sensing cells (statocytes) is a likely trigger of subsequent intracellular signalling (Morita, 2010). However, several studies have demonstrated that starch is important but not essential for gravity sensing (Strohm et al., 2011; Wolverton et al., 2011). The receptor that responds to gravity has not yet been identified. In the process of gravity perception, changes in the gravity vector are transduced into multiple intracellular signals [i.e. cytosolic pH, inositol 1,4,5-triphosphate (InsP3), and cytosolic calcium concentration] (Fasano et al., 2001; Plieth et al., 2002; Perera et al., 2006). The plant hormone auxin was identified as one of these signalling molecules, and its redistribution is polar. Auxin has been thought to be involved in gravitropic responses, based on the Cholodny–Went theory. Auxins were the first group of plant growth regulatory substances to be discovered (Went, 1928). Indole-3-acetic acid (IAA) was the first native representative of the group that was identified. Auxins are known to be involved in the regulation of basic growth processes, such as cell division and cell elongation at the tissue, organ, and whole-plant levels. The auxin molecule functions as a mobile signal between cells, tissues, and organs, and is involved in the spatial and temporal coordination of plant morphogenesis and responses to the environment in planta. Auxin is not only a mobile molecule; it also participates in downstream signalling. Many developmental processes appear to depend on the local asymmetric distribution of auxin molecules (Tanaka et al., 2006). These include embryo development and apical–basal axis formation in Arabidopsis thaliana (Friml et al., 2003), pattern formation and root development (Blilou et al., 2005), organ formation (Benkova et al., 2003), and phototropism and gravitropism (Blakeslee et al., 2004).
In addition to auxin, the phytohormone ethylene has been shown to be involved in the gravity response in plants (Madlung et al., 1999). In tomatoes, root penetration into the soil required cross-talk between ethylene and auxin (Santisree et al., 2011). A mutation of the Arabidopsis gene ARG1, which is involved in root gravitropism, showed resistance to ethylene but increased sensitivity to auxin (Sedbrook et al., 1998). The AGR gene encodes a membrane protein that is homologous with bacterial transporter proteins (Utsuno et al., 1998) as a polar auxin transporter (Chen et al., 1998). Another Arabidopsis mutant, clg1, showed resistance to ethylene in root gravitropism (Ferrari et al., 2000). In Arabidopsis, flavonoid accumulation by ethylene was found to be involved in root gravitropism (Buer et al., 2006). Additionally, the Arabidopsis mutant rha1 showed resistance to ethylene in roots and was found to be involved in root gravitropism (Fortunati et al., 2008). In six plant species tested, an ethylene biosynthesis inhibitor inhibited root curvature (Hoson et al., 1996). In Arabidopsis, a recent study showed that ethylene and gravity can affect root skewing and waving (Oliva and Dunand, 2007). The Arabidopsis mutant alh1 was shown to link the cross-talk between ethylene and auxin in the gravity response in roots (Vandenbussche et al., 2003). Moreover, acs6 and acs9 mutants showed defective hypocotyl length in the gravity response (Tsuchisaka et al., 2009). However, it is still controversial whether ethylene plays a positive or negative role in modulating the gravity response (Philosoph-Hadas et al., 1996; Buer et al., 2006).
Ethylene is a plant hormone involved in many plant growth and developmental processes, including seed germination, leaf and flower senescence and abscission, cell elongation, fruit ripening, nodulation, and responses to a wide variety of stressors (Crocker and Knight, 1908; Wang et al., 2002; Yoo et al., 2009). The biosynthesis of ethylene has been well documented in plants (Yang and Hoffman, 1984; McClellan and Chang, 2008). Ethylene is derived from the amino acid methionine, which is converted to S-adenosylmethionine (AdoMet) by S-adenosylmethionine synthetase. AdoMet is then converted to 1-aminocyclopropane-1-carboxylic acid (ACC) and 5′-deoxy-5′methylthioadenosine (MTA) by the enzyme 1-aminocyclopropane-1-carboxylase synthase (ACS) (Adams and Yang, 1979; Lin et al., 2009), which is the rate-limiting step in ethylene biosynthesis. MTA is recycled to methionine through the Yang cycle, which allows high rates of ethylene production without depleting endogenous methionine. ACC is converted to ethylene, CO2, and cyanide by ACC oxidase (ACO). The cyanide produced by this reaction is detoxified into β-cyanoalanine by the enzyme β-cyanoalanine synthase, preventing toxicity to plants under conditions of high ethylene biosynthesis.
ACS proteins play an important role in the ethylene biosynthesis pathway. The enzyme catalyses the conversion of AdoMet to ACC, and this reaction requires pyridoxal-5′-phosphate (PLP) as a cofactor. In most plant species, ACS is encoded by a multigene family that is regulated by various environmental and developmental factors (i.e. cytokinin, auxin, root hair development, fruit ripening, wounding, and pathogens). Arabidopsis has eight genes that encode active ACS proteins and an additional gene that encodes a catalytically inactive enzyme, ACS1 (Liang et al., 1992). The Arabidopsis ACS2, ACS6, ACS7, and ACS9 genes can be induced by hypoxia (Peng et al., 2005). Zhang’s group found that ACS2, ACS6, ACS7, ACS8, and ACS11 were involved in Botrytis cinerea-induced ethylene biosynthesis in Arabidopsis (Li et al., 2012). Based on the C-terminal sequences, ACS proteins in Arabidopsis can be divided into three main types (Chae and Kieber, 2005). Type I proteins have an extended C-terminus that contains three conserved serine residues that are targets for phosphorylation by mitogen-activated protein kinase 6 (MPK6) (Liu and Zhang, 2004) and a conserved serine residue that is a phosphorylation site for calcium-dependent protein kinase (CDPK; Tatsuki and Mori, 2001; Sebastià et al., 2004). Type II proteins have a shorter C-terminus that has only the CDPK phosphorylation site. Type III proteins have a very short C-terminal extension that lacks both phosphorylation sites. ACS proteins can act as homo- or heterodimeric proteins, similar to other PLP-dependent enzymes, and their ability to form active heterodimers might act to increase the versatility of ethylene responses (Tsuchisaka and Theologis, 2004), which enhances the ability to regulate ethylene production after exposure to different developmental and environmental stimuli.
Details on ACS turnover have been derived from studies of Arabidopsis ethylene-overproducing (Eto) mutants (Chae and Kieber, 2005). The eto mutants produce 10- to 40-fold more ethylene in the dark compared with the wild-type (WT) seedlings, and adopt a triple-response morphology (i.e. a morphology that etiolated seedlings adopt in the presence of ethylene) in the absence of exogenous ethylene application (Guzman and Ecker, 1990; Kieber et al., 1993). The cloning of ETO1 revealed that it encodes an E3 ligase component, a BTB/TPR protein. ETO1 binds to type II ACS proteins but not type I or type III ACS proteins (Wang et al., 2004; Yoshida et al., 2005, 2006). The disruption of ETO1 resulted in increased stability of the type II ACS protein ACS5 (Chae et al., 2003) and consequently increased ethylene biosynthesis. The stability of ACS proteins is also regulated by protein phosphorylation. Treatment of tomato cells with the protein kinase inhibitors K-252a and staurosporine inhibited the elicitor-dependent induction of ACS and ethylene biosynthesis (Grosskopf et al., 1990; Felix et al., 1991) through a mechanism that most probably involves increased turnover of the ACS protein (Spanu et al., 1994). In tomato cells, ACS2 was shown to be phosphorylated by CDPK from extracts of wounded tomato fruit (Tatsuki and Mori, 2001). The protein stability of ACS2 was found to be regulated by CDPK phosphorylation (Kamiyoshihara et al., 2010). The target of CDPK phosphorylation was the conserved serine residue Ser460 at the C-terminal region of the ACS protein. A novel CDPK phosphorylation motif was identified in the C-terminal domain of type II ACS proteins (Sebastià et al., 2004). The current model proposes that the phosphorylation of type I and type II ACS proteins blocks the ability of ETO1/EOL proteins to bind and inhibit the ubiquitination of these ACS proteins for their degradation by the 26S proteasome (Wang et al., 2004; Chae and Kieber, 2005).
The regulation of ethylene biosynthesis and ACS stability are also controlled by mitogen-activated protein kinases (MAPKs). In tobacco, a stress-induced MAPK (SIPK) is involved in the response to different stressors, including pathogen- and ozone-induced ethylene biosynthesis. The closest homologue of SIPK in Arabidopsis is MPK6. Therefore, MPK6 was used in an in vitro kinase assay. The results showed that MPK6 can phosphorylate ACS2 and ACS6 in vitro, and transgenic plants that overexpress a phosphomimic mutant of ACS6 showed increased ethylene production (Liu and Zhang, 2004). These results indicate that a pathway similar to the SIPK pathway in tobacco operates in Arabidopsis and that MPK6 phosphorylates ACS proteins, thereby decreasing their turnover and increasing ethylene biosynthesis after pathogen stress. A possible CDPK- and MPK6-regulated pathway was recently proposed by Ludwig et al. (2005). These findings highlight the complexity of phosphorylation-regulated signalling and ethylene biosynthesis in plants in response to different stressors.
Calcium is a ubiquitous secondary messenger in eukaryotic cells. In plants, intracellular calcium levels can modulate many growth and developmental processes, including plant hormones, light, gravity, and biotic and abiotic stress (Batistic and Kudla, 2012). Unlike most other ions, calcium does not freely diffuse within cells (Trewavas, 1999). Plants have multiple calcium stores, including in apoplasts, vacuoles, the nuclear envelope, the endoplasmic reticulum (ER), chloroplasts, and mitochondria. Different stimuli can trigger calcium efflux from specific organelles. After calcium is released, different calcium sensors that have an EF-hand motif that can specifically bind calcium [i.e. the EF-hand-containing proteins calmodulin (CaM) and CDPK] can recognize specific calcium signals in specific places and transduce them into downstream effects, including altered protein phosphorylation and gene expression patterns (Sanders et al., 1999, 2002; Dodd et al., 2010; Perochon et al., 2011; Liese and Romeis, 2012).
In Arabidopsis, the C-terminal domain in type I and type II ACS proteins can be phosphorylated by CDPK and MAPK. This phosphorylation of type I and type II ACS proteins blocks the ability of ETO1/EOL proteins to bind, thus inhibiting the ubiquitination of these ACS proteins and their degradation by the 26S proteasome. The degradation of type III ACS7 was recently found to also be mediated by the 26S proteasome (Lyzenga et al., 2012), but how ACS7 protein activity and stability are regulated is still unknown. The present study investigated whether the type III ACS protein ACS7 is phosphorylated by CDPK in vitro. An in vitro kinase assay was conducted to determine the phosphorylation of ACS7 by CDPK and identify the phosphorylation site in ACS7. Additionally, the protein–protein interaction between ACS7 and 14-3-3ω was confirmed using two independent methods. A T-DNA insertion knockout mutant, acs7-1, was also identified and used to investigate the previously uncharacterized functions of ACS7 in Arabidopsis. It was found that ACS7 is involved in root gravitropism in Arabidopsis.
Materials and methods
Plant growth conditions
Seeds from WT Arabidopsis thaliana plants (Columbia and Wassilewskija ecotype) were sterilized with chlorine for 3h with 100ml of 6% bleach and 3ml of 10 N sulphuric acid (H2SO4) and then spread onto plates that contained half-strength Murashige and Skoog medium and 0.5% sucrose. The seeds were placed in the dark for 2 d at 4 °C and then incubated in growth chambers for 16h with 100 μmol photon m–2 s–1 light and 8h in the dark at 23 °C. After seed germination, all of the plants were transferred to a 9F walk-in growth chamber under a short-day photoperiod condition (8h light/16h dark) or a long-day photoperiod condition (16h light/8h dark).
Preparation of E. coli DH5α competent cells for transformation
Escherichia coli competent cells were prepared. A single colony of DH5α was inoculated into 5ml of LB medium (1% tryptone, 0.5% yeast extract, and 1% NaCl) with shaking at 37 °C for 16h. The cells were then added to a 1 litre flask that contained 500ml of Super Optimal Broth medium with shaking at 37 °C for 3h. The cells were centrifuged at 2700 g (Beckman Coulter J2-MC, Brea, CA, USA) for 10min at 4 °C and gently resuspended in 130ml of 0.1M CaCl2 solution. The cells were then centrifuged at 2700 g for 10min at 4 °C, and the supernatant was removed. Finally, the cells were resuspended in 4ml of TB buffer and separated in a 1.5ml tube. DH5α competent cells were placed on ice, and 1 μl of plasmid was added to thaw the cells. The cells were placed on ice for 30min and heat-shocked for 90 s at 42 °C. The cells were then placed on ice for 2min. LB medium (1ml) was added to let the cells recover at 37 °C for 30min. After recovery, the cells were spread on plates and incubated overnight at 37 °C.
Purification of glutathione S-transferase (GST)-tagged protein
Escherichia coli was incubated in 40ml of 2× YT medium with 200 μg of ampicillin at 37 °C for 16–18h overnight, and 400ml of 2× YT medium was then added at 28 °C for 3h. After incubation, 220 μl of isopropyl β-d-1-thiogalactopyranoside (IPTG; 1M stock, 230mg ml–1) was added to a final concentration of 0.5mM and incubated for 3h at 37 °C. The cells were centrifuged at 6000rpm (Beckman Coulter J2-MC) for 30min at 4 °C. The supernatant was discarded, and 20ml of lysis buffer was then added for 15min. The cells were transferred to a 50ml Falcon tube and stored at –80 °C. The cells were incubated in water (~40 °C), and a sonicator (Misonix XL2020, Farmingdale, NY, USA) was used to break the cells. After sonication, the cells lysate was centrifuged at 10 000rpm for 30min at 4 °C. The supernatant was then transferred to a new 50ml Falcon tube. GST beads (1ml) were added and washed with GST binding buffer three times, and the solution was shaken for 1–3h in a cold room. The GST beads were centrifuged, and 10ml of GST binding buffer was added to wash the beads three times. TRIS buffer (10ml, 50mM, pH 7.5) was added to wash the beads, and the solution was transferred to a biospin column (Bio-Rad, Hercules, CA, USA) to allow the GST beads to adhere to the column. TRIS buffer (1.5ml, 50mM, pH 8.0) that contained 10mM glutathione (30mg per 10ml) was used to elute the protein, and the eluate was collected by a centrifugal filter (Amicon Ultra 10K, Millipore, Billerica, MA, USA).
6His-SUMO–ACS7 recombinant protein purification
6His-SUMO–ACS7 recombinant protein was incubated in 40ml of LB medium that contained 50 μg ml–1 ampicillin and shaken at 37 °C overnight. The overnight culture was inoculated with 400ml of LB medium with shaking at 37 °C. The cells were raised to an absorbance of optical density (OD) 0.4–0.6 (mid-log phase) at 600nm and then induced with 0.5mM IPTG and allowed to continue to grow for 3h. The cells were then harvested by centrifugation at 5520 g (Beckman Coulter J2-MC) for 30min. The cell pellet was resuspended in lysis buffer and stored at –80 °C. The cells were thawed in 40 °C water, sonicated, and centrifuged at 20 400 g for 30min (Beckman Coulter J2-MC). The supernatant was incubated with pre-washed Ni-NTA resin (GE) for 2h at 4 °C and washed with a first wash buffer and second wash buffer. 6His protein was eluted with 1.5ml of elution buffer. The eluate was collected with a centrifugal filter (Amicon Ultra 10K, Millipore) for buffer exchange. The fusion protein was resuspended in phosphate-buffered saline (PBS) buffer. Plasmid maps are shown in Supplementary Fig. S1 available at JXB online.
Protein quantification using the Bradford assay
The protein concentration measurement was based on the Bradford method using Protein Assay Dye (Bradford, 1976; catalog no. 500-0006, Bio-Rad). Protein assay dye (100 μl) was mixed with 900 μl of dH2O and added to different concentrations of bovine serum albumin to reconstitute the standard curve. Sample absorbance was read at 595nm.
Fusion peptide design and construction
Approximately 50–60 nucleotide long forward and reverse primers were used to self-ligate in a temperature gradient and had a sticky-end AscI and BamHI restriction enzyme recognition site. After phosphorylation by polynucleotide kinase at the 5′ end for 30min, the double-stranded primer was constructed into an NRV vector and transformed into BL21 for fusion protein expression.
In vitro kinase assay
The in vitro kinase assay was performed according to a modified method described previously (Curran et al., 2011). ATP (50 μM, spiked with 1.25 μCi of [γ-32P]ATP) was added to begin the kinase reaction at a final volume of 10 μl, which consisted of 300ng of purified CDPK, 3 μg of fusion protein substrate, and standard kinase reaction buffer. The reactions were incubated for 15min at room temperature and stopped by SDS sample buffer. All of the samples were loaded onto a 12% SDS–PAGE loading well for electrophoresis, and labelling signals were normalized to the amount of protein determined from Coomassie Brilliant Blue-stained gels after running SDS–PAGE.
Protoplast isolation
Arabidopsis protoplasts were isolated according to a modified method described previously (Yoo et al., 2007). The leaves from 4-week-old plants were excised and subjected to an enzyme solution for 2h at room temperature. The enzyme solution that contained protoplasts was filtered with a miracloth and centrifuged at 100 g (Kubota 2420, Japan) to pellet the protoplasts in a 15ml tube for 3min. The supernatant was removed, and the protoplasts were washed three times in W5 solution. The protoplasts were resuspended in Mmg solution with 2.5×105 protoplasts in 1ml before polyethylene glycol (PEG)-mediated transformation.
Plasmid construction and transformation for transient expression
The open reading frame (ORF) of AtACS7 was amplified using designated primers from cDNA. The amplified ORF was inserted into the p2YGW7 vector (Invitrogen). The method of transiently expressed plasmid transformation was performed as previously described (Yoo et al., 2007). Plasmids (10 μg) and 200 μl of protoplasts were added to a 15ml tube and gently mixed. PEG solution (200 μl) was then added and incubated at room temperature for 10min. The PEG solution that contained protoplasts was diluted with 1ml of W5 solution and gently mixed. Protoplasts were centrifuged at 100 g (Kubota 2420) to pellet the protoplasts for 3min. The supernatant was removed, and the protoplasts were washed twice with W5 solution. The protoplasts were resuspended with 1ml of W5 solution in each well of a 6-well tissue culture plate and incubated at room temperature. After 12–16h, yellow fluorescent protein (YFP) fluorescence was detected with a confocal microscope (SP5, Leica, Microsystems, Germany). Plasmid maps are shown in Supplementary Fig. S1 at JXB online.
Plasmid construction of bimolecular fluorescence complementation (BiFC) and transformation of plasmids for BiFC analysis
BiFC analyses were performed according to a modified method described previously (Yoo et al., 2007; Lee et al., 2012). The ORF of AtACS7 was amplified using designated primers from cDNA. The amplified ORF was inserted into the pEarleyGate201-YN vector or pEarleyGate202-YC driven by the 35S promoter and fused to YFP-N or YFP-C in-frame. Plasmids (10 μg; YFP-N and YFP-C) and 200 μl of protoplasts were added to a 15ml round-bottomed tube and gently mixed. PEG solution (110 μl) was added and incubated at room temperature for 10min. The PEG solution that contained protoplasts was diluted with 550 μl of W5 solution and gently mixed. Protoplasts were centrifuged at 100 g (Kubota 2420) to pellet the protoplasts for 3min. The supernatant was removed, and the protoplasts were washed twice with W5 solution. Protoplasts were resuspended with 1ml of W5 solution in each well of a 6-well tissue culture plate and incubated at room temperature. After 12–16h, YFP fluorescence was detected by a confocal microscope. Plasmid maps are shown in Supplementary Fig. S1 at JXB online.
Site-directed mutagenesis
Site-directed mutagenesis was performed using the QuikChange Lightning kit (Stratagene, La Jolla, CA, USA). Two complementary oligonucleotides that contained the desired mutation, flanked by an unmodified nucleotide sequence, were designed. Mutated nucleotides were amplified by PCR. The DpnI restriction enzyme (2 μl) was then directly added to each amplification reaction and incubated at 37 °C for 5min to digest the parental supercoiled double-stranded DNA. The DNA treated with 2 μl of DpnI was transformed into the DH5α competent cells.
SDS–PAGE
SDS–PAGE was performed according to a modified method described previously (Laemmli, 1970). A gel preparation system (Bio-Rad) was used to prepare a 4% stacking gel and 12% resolving gel. The samples were supplemented with sample buffer, heated at 95 °C for 5min, and subjected to SDS–PAGE at a constant voltage of 100V until the protein dye left the gel.
Quartz crystal microbalance (QCM) sensor washing and analysis
QCM was performed according to a modified method described previously (Matsunaga and Ueda, 2010). A volume of 500 μl of 1% SDS was added to the sensor and left for 3min. Double-distilled H2O (ddH2O) was used to wash the sensor. Piranha solution (3 μl; 99% H2SO4:30% H2O2, 3:1) was added to the sensor and left for 5min. ddH2O was used to wash the sensor. This procedure was repeated twice. The sensor was placed into an AFFINIX QN μ (INITIUM, Japan), and the basic frequency was measured. A volume of 500 μl of PBS buffer was added to the sensor, and the frequency was left to stabilize. 6His-SUMO–ACS7 protein was added to the sensor until the sensor coating was saturated. The sensor was washed twice with PBS buffer, and 500 μl of PBS was added to the sensor. 14-3-3ω protein was added to the sensor, and the dissociation constant K d was measured.
Genomic DNA extraction
The leaves of 3-week-old plants were excised and placed into a 1.5ml tube with liquid nitrogen. The leaves were ground into a powder, and 750 μl of Genomic DNA extraction buffer and 50 μl of 20% SDS were added. The 1.5ml tube was heated at 65 °C for 10min. After heating, 250 μl of 5M potassium acetate was added and placed on ice for 10min. Chloroform (200 μl) was added into the 1.5ml tube and centrifuged for 10min at 13 000rpm. A total of 800 μl of the supernatant was moved to a new 1.5ml tube. Isopropanol (560 μl) was added and centrifuged at 13 000rpm for 10min. The pellet was washed with 70% ethanol and resuspended in ddH2O. Genomic DNA was used to determine the homozygosity of the T-DNA insertion in the acs7-1 mutant using PCR.
RNA extraction
RNA was isolated using REzol™ C&T reagent (Protech, Taipei, Taiwan) and converted to cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA). The leaf sample was ground into a powder with liquid nitrogen, and 1ml of REzol™ C&T and 200 μl of chloroform were added. The sample was centrifuged at 13 000rpm for 15min and moved to a new 1.5ml tube. Isopropanol (500 μl) was added and centrifuged at 13 000rpm (Sigma 1-15K, St Louis, MO, USA) for 10min. The pellet was washed with 75% ethanol and resolved by diethylpyrocarbonte (DEPC)-treated H2O. Total RNA (2 μg) was subjected to cDNA synthesis. The same amount of cDNA was used for PCR analysis.
Root curvature and gravity-sustaining response ratio measurement
The root gravity response was measured as previously described (Sukumar et al., 2009) (Supplementary Fig. S2 at JXB online). Three-day-old seedlings were transferred to either control agar (0.8%) or agar supplemented with multiple chemicals [i.e. EGTA (Sigma), LaCl3, LiCl, ACC, 1-naphthylphthalamic acid (NPA), and ruthenium red (RR)] at the indicated concentrations. After 12–24h of vertical growth, the plates were rotated 90 ° counterclockwise. Photographs of the plants were taken at specific time points (Time 0) after reorientation using a digital camera. The root tip curvature (in degrees) after reorientation (the angle difference of before and after reorientation) was measured every 12h by ImageJ software, and the gravity-sustaining response ratio was determined using the percentage of roots that grew toward the direction of the new gravity vector.
Results
AtACS7 can be phosphorylated by AtCDPK16 in vitro
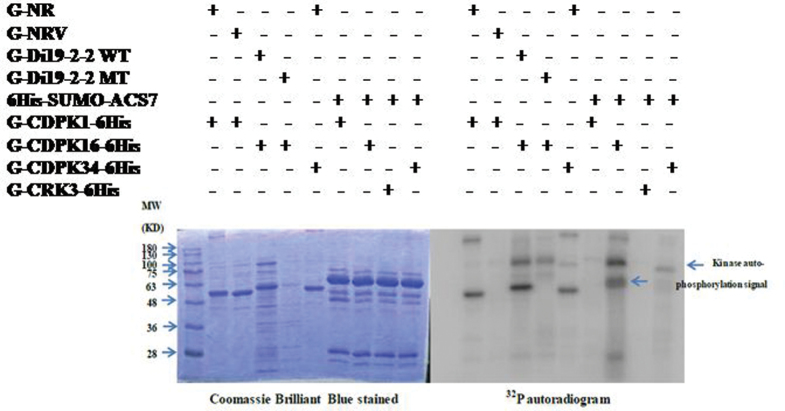
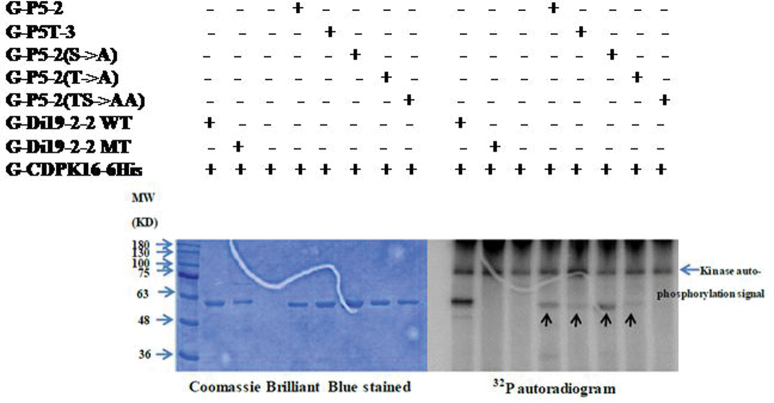
A recent study found that type I and type II ACS proteins can be CDPK substrates (Sebastià et al., 2004). Although the type III ACC synthase ACS7 has a shorter C-terminal domain and does not have a predicted CDPK phosphorylation site, ACS7 may still be a substrate of CDPK. AtCDPK1, AtCDPK16, AtCRK3, and AtCDPK34 were used for the in vitro kinase assay against the full-length ACS7 recombinant protein 6His-SUMO–AtACS7. G-NR, G-Di19-2-2 WT, and G-Di19-2-2 mutant (MT) fusion peptides were used as controls for the kinase assay as described previously (Curran et al., 2011). The kinase assay results indicated that CDPK16 can phosphorylate the recombinant protein 6His-SUMO–AtACS7 in vitro (Fig. 1).
Fig. 1.
SUMO–AtACS7 was phosphorylated by AtCDPK16 in vitro. Recombinant protein 6His-SUMO–ACS7 was used as a substrate, and four kinds of recombinant kinases (G-AtCDPK1-6His, G-AtCDPK16-6His, G-AtCRK3-6His, and G-AtCDPK34-6His) were used to perform the kinase assay in vitro. The result indicates that 6His-SUMO–AtACS7 can only be phosphorylated by G-AtCDPK16-6His in vitro. G-NR is a fusion protein, with GST fused with a peptide of nitrate reductase (TLKRTASTPFM), and this peptide is known to be recognized by G-AtCDPK1-6His; G-NRV is a vector-only protein; G-Di19-2-2WT is also a fusion protein in which GST is fused with a peptide (DVLKSEQKEMSYREDPY); this peptide can be recognized by G-AtCDPK16-6His, and G-Di19-2-2MT is similar to G-Di19-2-2WT but with serine mutated to alanine. The molecular weights of all the fusion proteins are ~55kDa and that of SUMO–ACS7 is ~63kDa; the peptide phosphorylation signal is marked with a arrowhead, and the kinase autophosphorylation signal is ~100kDa (right panel).
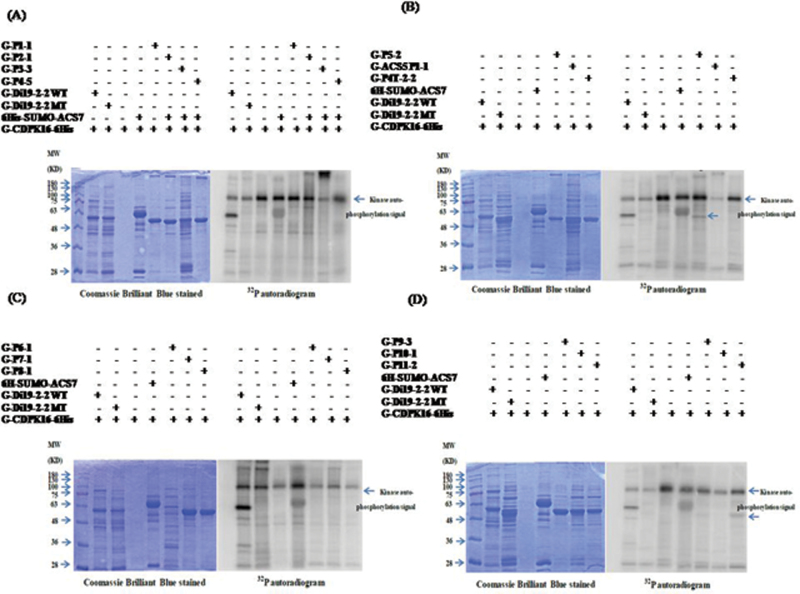
The possible phosphorylation sites in ACS7 were further investigated. Liquid chromatography–mass spectrometry/mass spectrometry (LC-MS/MS) was used to identify the peptides in the ACS7 recombinant protein phosphorylated by CDPK. According to the LC-MS/MS results and conserved Ser/Thr residues of ACS in different plant species, 13 fusion peptides that contained possible phosphorylation sites were designed as candidates for the in vitro kinase assays (Supplementary Table S1 at JXB online). After the kinase assay, it was found that the fusion peptides P5-2, which contains the peptide V(294)GT(296)IYS(299)YNDNV(304), and P11-2, which contains the peptide V(207)RGVLIT(213)NPS(216)NPL(219), were labelled in the 32P autoradiogram (Fig. 2). This result indicates that P5-2 and P11-2 can be phosphorylated by CDPK16 in vitro.
Fig. 2.
Peptide P5-2 and P11-2 were phosphorylated by CDPK16 in vitro. The fusion proteins (GST fused with small peptides) were designed to perform the kinase assay and determine which serine or threonine can be phosphorylated by G-CDPK16-6His. P5-2 contains the peptide V(294)GTIYSYNDNV(304); and P11-2 contains the peptide V(207)RGVLITNPS(216)NPL(219). G-Di19-2-2WT phosphorylated by G-CDPK16-6His is a positive control; G-Di19-2-2MT is a negative control; and 6His-SUMO–ACS7 is a full-length ACS phosphorylation control. The results indicate that P5-2 (in B) and P11-2 (in D) can be phosphorylated by CDPK16. The molecular weight of the fusion peptide is ~55kDa; peptide phosphorylation signals are marked with arrowheads, and the kinase autophosphorylation signal is ~100kDa (right panels).
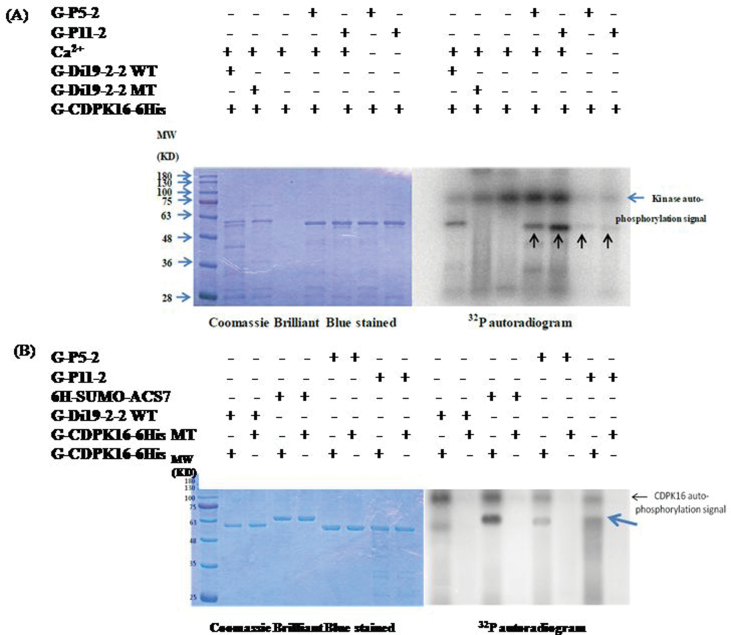
A previous study showed that CDPK phosphorylates substrates in a calcium-dependent manner (Hetherington and Trewavas, 1984). Because the fusion peptides P5-2 (VGTIYSYNDNV) and P11-2 (VRGVLITNPSNPL) were shown to be phosphorylated by CDPK16, experiments were carried out to determine whether this phosphorylation is calcium dependent. Figure 3A shows that when calcium ions were added in the kinase reaction buffer, P5-2 and P11-2 phosphorylation signals were detected. When calcium ions were depleted in the reaction buffer, phosphorylation was abolished. This confirms that P5-2 and P11-2 phosphorylation is calcium dependent.
Fig. 3.
The phosphorylation of P5-2 and P11-2 is calcium dependent. (A) In order to confirm that this phosphorylation reaction is calcium dependent, buffer with or without calcium ions was tested. The result indicated that the substrate kinase phosphorylation reaction is calcium dependent (lanes 4–7). The amino acid sequences in P5-2 and P11-2 are VGTIYSYNDNV and VRGVLITNPSNPL. Peptide phosphorylation signals are marked with arrowheads. (B) An in vitro kinase assay was carried out using mutated G-CDPK16-6His (G-CDPK16-6His MT) having double mutations on both Ser274 and Ser541. Both serines were mutated to alanine. The AtACS7 fusion protein and peptide phosphorylation signals are marked with arrowheads.
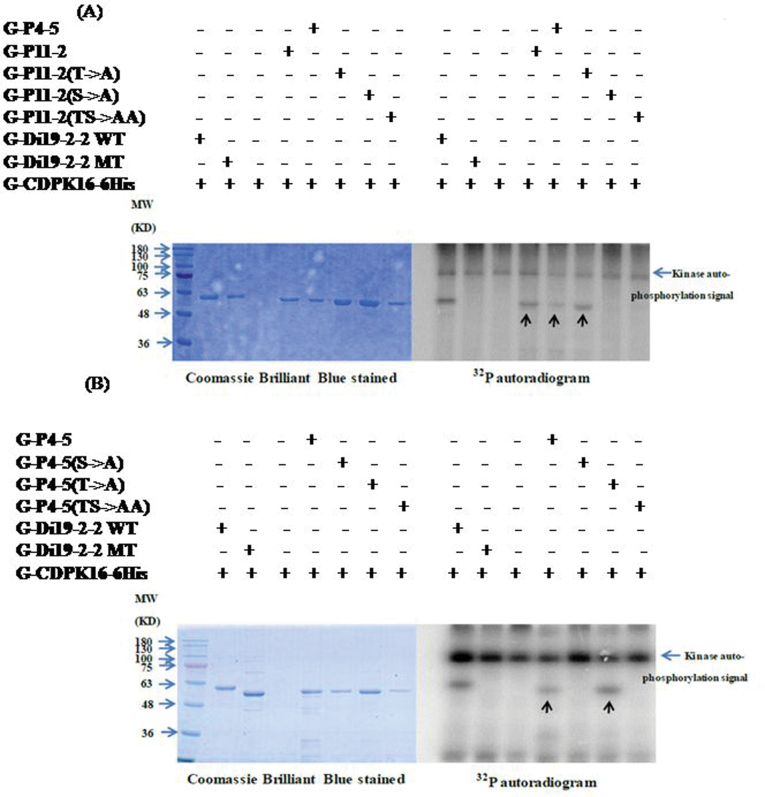
To confirm that the in vitro phosphorylation of AtACS7 fusion peptides was due to AtCDPK16 activity, a control experiment was carried out. Two autophosphorylation sites (Ser274 and Ser541) of the AtCDPK16 recombinant fusion protein (Hegeman et al., 2006) were mutated by site-directed mutagenesis. The mutated AtCDPK16 recombinant fusion protein (G-CDPK16-6His MT) was used in the in vitro kinase assay against AtACS7 fusion peptides. The results showed that the autophosphorylation of the mutated recombinant protein (G-CDPK16-6His MT) was greatly reduced (Fig. 3B; Supplementary Fig. S3 at JXB online). In addition, the phosphorylation of the AtACS7 fusion peptides was greatly reduced (Fig. 3B; Supplementary Fig. S3). This indicated that the phosphorylation of AtACS7 fusion peptides in vitro resulted from AtCDPK16 but not from other bacterial kinases. The P5-2 and P11-2 peptides each contain one serine and one threonine residue. Furhter experiments were carried out to determine which amino acid is the target for CDPK16 or whether both can be phosphorylated by CDPK16. To address this issue, a point mutation was created in the two peptides using site-directed mutation of either serine or threonine to alanine (Supplementary Table S1 at JXB online). For P11-2, the results indicated that only Ser216 was recognized by CDPK16 (Fig. 4). To confirm the phosphorylation sites, the fusion peptide P4-5, which has 10 amino acids that overlap with P11-2, was used for the kinase assay. A point mutation of either serine or threonine to alanine was designed to verify the phosphorylation site in P11-2. The results showed that only Ser216 was recognized by CDPK16 (Fig. 4B). This result was consistent with the results presented in Fig. 4A. The serine residue (Ser216) in both P11-2 and P4-5 was shown to be an AtCDPK16 phosphorylation site.
Fig. 4.
Phosphorylation of Ser216 was confirmed by site-directed mutagenesis. In order to identify the phosphorylation residue in P11-2, site-directed mutagenesis was performed. P4-5 is a peptide which has a 10 amino acid sequence overlapping with P11-2. P11-2 S→A is a serine to alanine mutation peptide in P11-2; P11-2 T→A is a threonine to alanine mutation peptide; P11-2 TS→AA is a double mutation peptide of serine and threonine to alanine. The results indicated that only serine (Ser216) but not threonine (Thr213) in P11-2 can be phosphorylated by G-CDPK16-6His in vitro. Peptide phosphorylation signals are marked with arrowheads.
For P5-2, a fusion peptide with a single point mutation of either threonine or serine to alanine and double mutations of both serine and threonine to alanine were used to perform the kinase assay. The results showed that both Thr296 and Ser299 were recognized by CDPK16 (Fig. 5). Specifically, the phosphorylation of Ser299 was also supported by the MS/MS data (Supplementary Fig. S4 at JXB online). In conclusion, based on the in vitro assay, three AtCDPK16 phosphorylation sites, Ser216, Thr296, and Ser299, were identified. These results suggest that AtACS7 may be involved in calcium signalling.
Fig. 5.
Site-directed mutagenesis of the hosphorylation site on peptide P5-2. In order to identify the phosphorylation residue in P5-2, site-directed mutagenesis was performed. P5T-3 is a peptide which has a nine amino acid sequence overlapping with P5-2. P5-2 S→A is a peptide with a mutation of serine (Ser299) to alanine in P5-2; P5-2 T→A is a threonine (Thr296) to alanine mutation peptide; P5-2 TS→AA is a double mutation peptide of serine and threonine to alanine. The results indicated that both serine and threonine in P5-2 can be phosphorylated by G-CDPK16-6His in vitro. Peptide phosphorylation signals are marked with arrowheads.
The subcellular localization of AtACS7 is in the cytosol
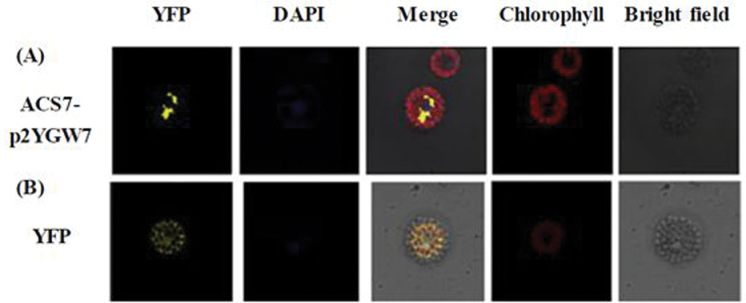
ACS proteins have been found to be cytosolic proteins (Yip et al., 1991). To confirm further whether AtACS7 is actually localized in the cytosol, transient expression of ACS7 protein in protoplasts was detected. The 35S::YFP:ACS7 plasmid was transferred to Arabidopsis protoplasts, and confocal microscopy was used to observe the localization of the recombinant YFP fused with ACS7. As expected, ACS7 protein was localized in the cytosol based on the transient assay (Fig. 6).
Fig. 6.
Subcellular localization of AtACS7 in Arabidopsis protoplasts in a transient expression assay. Wild-type Arabidopsis protoplasts were transfected with (A) 35S:YFP:AtACS7 and (B) 35S:YFP (control) constructs. YFP signals were observed by confocal microscopy. The blue signal showed a nucleus stained with 4′,6-diamidino-2-phenylindole (DAPI); the red signal showed chlorophyll with autofluorescence; and merge shows YFP, DAPI, chlorophyll, and bright field signals.
Protein–protein interaction between ACS7 and 14-3-3ω was confirmed using BiFC and QCM
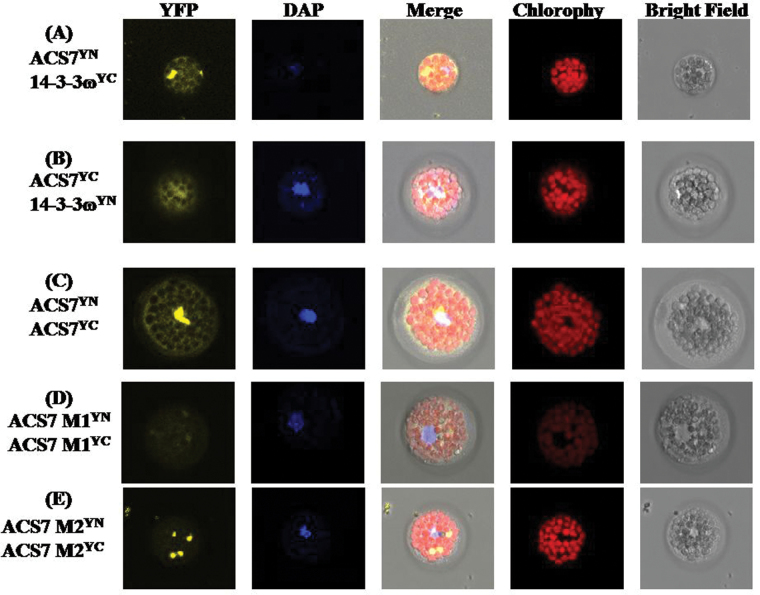
14-3-3 proteins have been regarded as scaffold proteins that can bind to phosphorylated proteins (Sehnke et al., 2002). A previous study showed that ACS7 can be a client of 14-3-3ω in Arabidopsis (Chang et al., 2009; Yoon and Kieber, 2013). Whether 14-3-3ω physically interacts with ACS7 and modulates its functions was investigated. Previous studies showed that ACS7 can form a homodimer to perform its functions (Tsuchisaka and Theologis, 2004), and fusion proteins ACS7-YN and ACS7-YC were used as a positive control (Supplementary Fig. S5B at JXB online). ACS7 formed homodimers as expected (Tsuchisaka and Theologis, 2004). Additionally, NR peptide, which contains a 14-3-3 interaction site, interacted with 14-3-3ω as expected. Based on the transient expression results, ACS7 physically interacted with 14-3-3ω in the cytosol (Supplementary Fig. S5E, F). Both ACS7-YN and ACS7-YC interacted with 14-3-3ω-YN.
To investigate the functional relevance of the identified phosphorylation sites, ACS7-YN and ACS7-YC, each with Ser216 point-mutated to alanine, were tested for interactions using BiFC. ACS7-YN and ACS7-YC, each with Thr296 and Ser299 double-mutated to alanine, were also tested. Both BiFC results showed that the mutation of these phosphorylation sites resulted in an altered subcellular localization pattern. The dimer formation signal was weaker and not evenly distributed in the entire cytoplasm as it was in the WT (Fig. 7C–E), suggesting that the phosphorylation of AtACS7 may have important functions.
Fig. 7.
Protein–protein interactions with ACS7. (A) Co-expression of NR-nEYFP and 14-3-3-cEYFP (positive control). (B) Co-expression of NR-cEYFP and 14-3-3-nEYFP (positive control). (C) Co-expression of ACS7-nEYFP and 14-3-3cEYFP. (D) Co-expression of ACS7-M1. (E) Co-expression of ACS7-M2. (D) and (E) are ACS7 mutant proteins for testing ACS7 homodimer formation. M1: Thr296 and Ser299 of AtACS7 were both mutated to alanine. M2: Ser216 of AtACS7 was point mutated to alanine. YFP signals were observed by confocal microscopy. The blue signal shows a nucleus stained with 4′,6-diamidino-2-phenylindole (DAPI); the red signal showed chlorophyll with autofluorescence; and merge shows YFP, DAPI, chlorophyll, and bright field signals.
To confirm further the protein–protein interaction between ACS7 and 14-3-3ω, QCM analysis was performed. The recombinant protein G-14-3-3ω was first coated on the sensor, and then 6His-SUMO–ACS7 protein was injected into the PBS buffer. If the recombinant protein 6His-SUMO–ACS7 can physically interact with G-14-3-3ω, then the sensor frequency would change. The results showed that 6His-SUMO–ACS7 interacted with G-14-3-3ω in vitro (Supplementary Fig. S6A at JXB online). The dissociation constant (K d) was 9.29±1.99×10–9 M (Supplementary Fig. S6C). A previous study showed that 14-3-3 is a scaffold protein that can interact with its clients that have already been phosphorylated (Wu et al., 1997). To determine whether the interaction between ACS7 and 14-3-3ω is phosphorylation dependent and whether the interaction is caused by ACS7 phosphorylation catalysed by CDPK16, an interaction assay between phosphorylated 6His-SUMO–ACS7 and G-14-3-3ω was performed. 6His-SUMO–ACS7 was phosphorylated by CDPK16 in vitro in advance. The results showed that phosphorylated 6His-SUMO–ACS7 can physically interact with G-14-3-3ω in vitro (Supplementary Fig. S6B), and the K d was 5.05±1.96×10–9 M (Supplementary Fig. S6D). The QCM results of tag-only controls did not show any interaction, as expected (Supplementary Fig. S6E, F). Altogether, the results are consistent with the previous discovery that ACS7 forms a protein complex with 14-3-3ω protein (Chang et al., 2009; Yoon and Kieber, 2013). The interaction between ACS7 and 14-3-3ω protein appeared to be both phosphorylation dependent and phosphorylation independent.
Identification of acs7-1 loss-of-function mutant line
To study the functions of AtACS7 in planta, an acs7-1 mutant line was ordered from the Arabidopsis Biological Research Center (http://abrc.osu.edu/). In this mutant line, T-DNA was inserted into the third exon of the AtACS7 gene (Supplementary Fig. S7A at JXB online). The background of the mutant line was Wassilewskija (WS). Genomic PCR was used to confirm the T-DNA insertion site. The T-DNA primer GBK5-F paired with the gene primer acs7-1-R, and the gene primer acs7-1-F paired with acs7-1-R were used to perform PCR. It was found that only the T-DNA primer GBK5-F with acs7-1-R flanked the DNA fragment, and acs7-1-F with acs7-1-R did not (Supplementary Fig. S7B). The results showed that the acs7-1 mutant was homozygous.
The acs7-1 mutant is less sensitive to inhibition by a calcium chelator and the channel blocker LiCl in the gravity response
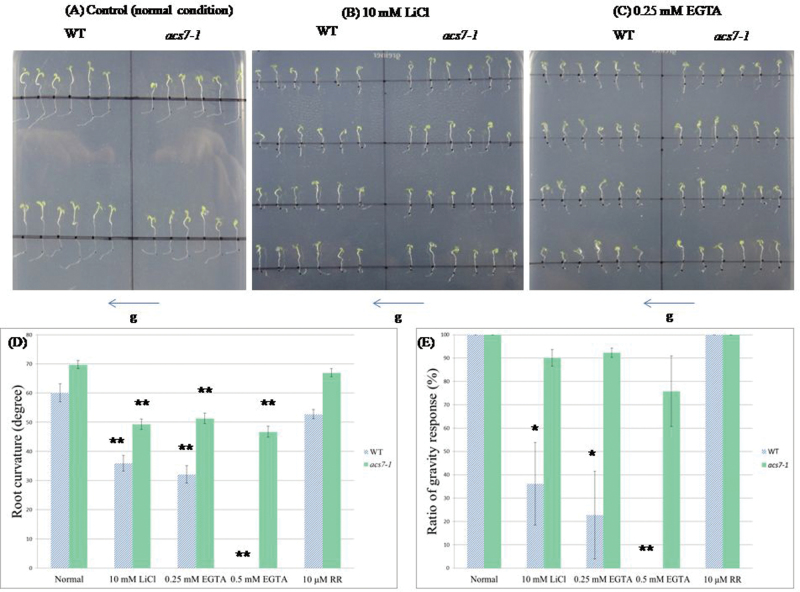
A previous study indicated that acs7-1 has multiple phenotypes (i.e. lower ethylene emission, larger cotyledons and true leaves, and longer primary root length; Dong et al., 2011). Additionally, the acs7-1 mutant exhibited hypersensitivity to abscisic acid (ABA) during seed germination (Dong et al., 2011). However, no studies of which the authors are aware have discussed the aspect of calcium in the acs7-1 mutant. The phenotype of the acs7-1 mutant line has been widely described (Tsuchisaka et al., 2009; Dong et al., 2011). The acs7-1 mutant exhibits early flowering, slightly reduced ethylene production, larger cotyledons, and enhanced salt tolerance. Because little is known about the relationship between ethylene biosynthesis and calcium (Raz and Fluhr, 1992; Philosoph-Hadas et al., 1996), several calcium channel blockers (i.e. LiCl, LaCl3, and RR), a calcium chelator (i.e. EGTA), and the CaM antagonist chlorpromazine (CPZ) were used to investigate the relationships between them. AtACS7 can be phosphorylated by CDPK in vitro, and it is hypothesized that AtACS7 may be involved in calcium signalling. Previous studies showed that calcium ions are important for the gravity response. When the WT seedlings are treated with calcium channel blockers or a calcium chelator, the plants may lose their gravity response (Hasenstein and Evans, 1988; Friedman et al., 1998; Toyota et al., 2008). To investigate the relationship between acs7-1 and calcium, the calcium channel blockers LiCl, LaCl3, and RR, the calcium chelators EGTA, and the CaM antagonist CPZ were used to determine whether calcium ions influence acs7-1. The results indicated that LaCl3 and CPZ did not cause significant differences between the WT and the acs7-1 mutant (Supplementary Figs S8, S9 at JXB online). Surprisingly, EGTA treatment caused a loss of the gravity response in the WT but not in the acs7-1 mutant. Figure 8C–E shows that acs7-1 can retain gravitropism, but the WT almost lost gravitropism. Moreover, 10mM LiCl treatment had the same effect as EGTA treatment (Fig. 8B, D, E). Collectively, the results indicate that ACS7 is involved in root gravitropism in a calcium-dependent manner in Arabidopsis.
Fig. 8.
Multiple calcium chelator and channel blockers affected the gravity response in acs7-1. The WT (WS) and the acs7-1 mutant were grown in half-strength Murashige and Skoog (1/2MS) medium for 3 d and transferred to 1/2MS medium which contained (A) control (normal conditions), (B) 10mM LiCl, (C) 0.25mM EGTA and other calcium channel blockers, and grown for 1 d. The gravity vector is downwards as the arrowhead indicates. The root curvature (degree) and ratio of gravity response (%) were measured in both the WT and the acs7-1 mutant after 24h (D and E). The average and SE are presented (n=60) for multiple calcium chelators or channel blockers in three independent experiments in (C), and the ratio of the gravity response is presented (n=60) for multiple calcium chelators or channel blockers in (D). RR, ruthenium red. Significant differences between normal conditions and treatment groups are indicated by * at P < 0.05 and by ** at P < 0.01 by Student’s t-test.
The acs7-1 mutant is less sensitive to ethylene in the triple response
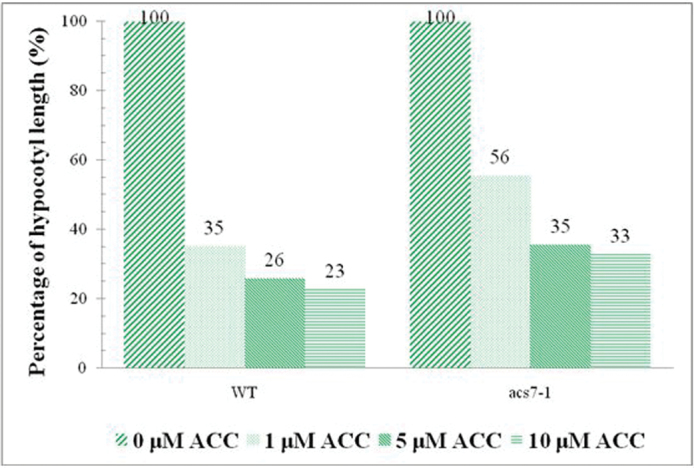
To determine whether the acs7-1 mutant displays ethylene-related phenotypes that are different from the WT in etiolated seedlings, the hypocotyl length was measured in the dark for 5 d and analysed by ImageJ software. The triple-response results showed that the acs7-1 mutant is less sensitive to ethylene compared with the WT (Fig. 9).
Fig. 9.
Triple response in theWT and acs7-1 mutant. The experiment was performed at 22°C for 5 d in the dark. Hypocotyl length and the percentage of hypocotyl length relative to normal conditions were measured by ImageJ. The average and SE are presented (n=60) for ACC in three independent experiments. A higher concentration of ACC in the dark shows reduced hypocotyl length more in the WT than in the acs7-1 mutant.
Gravity response under optimal ACC concentration
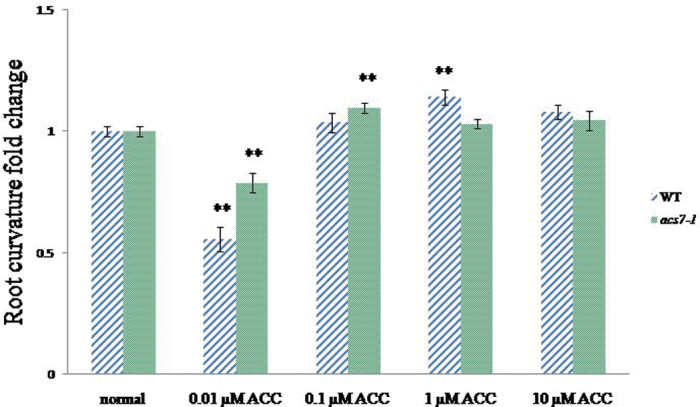
Ethylene has been suggested to regulate the gravity response. Some studies indicate that it plays a negative role in gravitropism (Buer et al., 2006), but other studies suggest a positive role in gravibending (Philosoph-Hadas et al., 1996). To investigate the effects of ethylene on gravitropism, gradient concentrations of ACC were introduced in the medium. Both the WT and the acs7-1 mutant responded to certain concentrations of ethylene that positively regulated the gravity response (WT at 1 μM ACC; acs7-1 at 0.1 μM ACC). A specific concentration of ethylene with 0.01 μM ACC negatively regulated the gravity response in both the WT and the acs7-1 mutant (Fig. 10).
Fig. 10.
ACC gradient for gravity response. The WT (WS) and acs7-1 mutant were grown in half-strength Murashige and Skoog medium for 3 d and transferred to medium which contained 0.01, 0.1, 1, or 10 μM ACC, and the root curvature of the root tip was measured by ImageJ after a 12h change of the gravity vector (turned counterclockwise by 90 °). Significant differences between normal conditions and treatment groups are indicated by ** at P < 0.01 by Student’s t-test.
The acs7-1 mutant is less sensitive to NPA-mediated inhibition of the gravity response
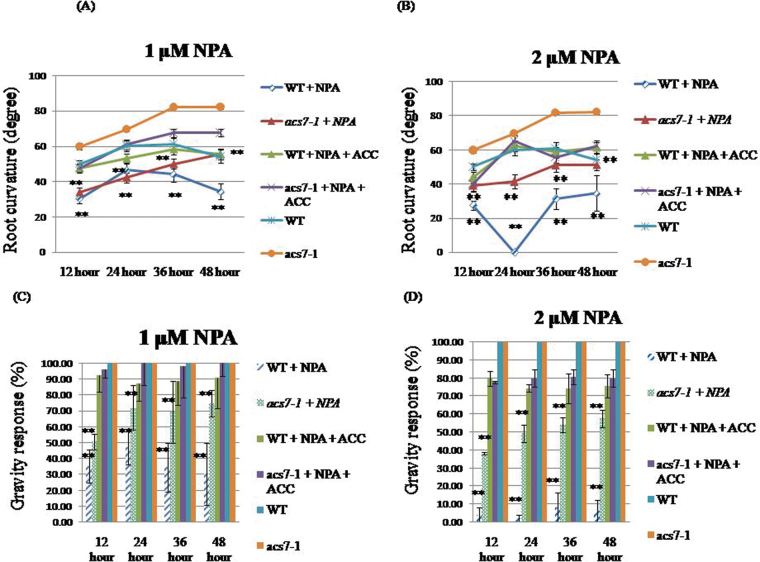
Some auxin polar transport inhibitors, including NPA and 2,3,5-triiodobenzoic acid (TIBA), were found to inhibit auxin polar transport. For example, TIBA can compete with auxins for translocation across the plasma membrane (Depta and Rubery, 1984), and NPA was found to disturb the polarity of cell division (Thomson et al., 1973). Therefore, NPA has long been used as an inhibitor of auxin polar transport (Thomson et al., 1973). The gravity response is greatly reduced by NPA treatment. To investigate the effects of NPA on the gravity response in the WT and the acs7-1 mutant, different concentrations (12 μM and 2 μM) of NPA were tested. The WT showed a smaller root curvature and lower gravity response than the acs7-1 mutant (Fig. 11A–D) in response to NPA treatment. This indicates that the acs7-1 mutant was less sensitive to the inhibitory effect of NPA. Exogenous ACC (0.1 μM) was added to the medium with NPA, which greatly reduced the effect of NPA and recovered root gravitropism (Fig. 11A–D). These results suggest that AtACS7 is involved in the root gravity response through an unknown cross-talk relationship between auxin and ethylene.
Fig. 11.
Root curvature and the ratio of the gravity response were affected by NPA in root tips. The WT (WS-4) and acs7-1 mutant were grown in half-strength Murashige and Skoog (1/2MS) medium for 3 d and transferred to medium which contained 1 μM or 2 μM NPA (A–D); the percentage gravity response was measured every 12h for 2 d. The percentages compared with normal conditions are presented (n=36) in three independent experiments. For ACC treatment, the WT (WS-4) and acs7-1 mutant were grown in 1/2MS medium for 3 d and transferred to medium which contained 1 μM or 2 μM NPA combined with 0.1 μM ACC (A and B); the root tip curvature was measured every 12h for 2 d by ImageJ software. The average is presented (n=36) in three independent experiments. The percentage gravity response was measured every 12h for 2 d. The percentage compared with normal conditions is presented (n=36) in three independent experiments (C and D). Significant differences between normal conditions and NPA treatment groups are indicated by ** at P < 0.01 by Student’s t-test.
Discussion
Figure 1 shows the phosphorylation of AtACS7 catalysed by AtCDPK16 in vitro. However, this result is different from the previous expectation that type III ACS proteins have no predicted CDPK phosphorylation sites and may not be a substrate of CDPKs. To date, only one Arabidopsis AtCDPK16 substrate, AtDi19-2, has been documented (Curran et al., 2011). The present study showed that AtCDPK16 can phosphorylate AtACS7 in vitro, but whether AtACS7 is an AtCDPK16 substrate requires further studies. Several consensus CDPK phosphorylation motifs are known (Harper and Harmon, 2005). The first consensus phosphorylation motif is ϕ-5-X-4-Basic-3-X-2-X-1-S, in which the underlined S is phosphorylated, X is any residue, and ϕ is a hydrophobic residue (Huang and Huber, 2001). The second consensus phosphorylation motif is Basic-9-Basic-8-X-7-Basic-6-ϕ-5-X-4-X-3-X-2-X-1-S-X+1-Basic+2. The third consensus phosphorylation motif is ϕ-3-R-2-ϕ-1-S-ϕ+1-x-K+3-R+4, which was defined as an ACS motif in a previous study (Sebastià et al., 2004). The identified phosphorylation site of AtACS7, Thr296, perfectly matches the first consensus phosphorylation motif. The phosphorylation motif S-X-R was recently identified from the AtCDPK16 substrate AtDi19-2 (Curran et al., 2011). In the present study, Ser216 and Ser299 were found to be phosphorylated by CDPK16 in ACS7 in vitro (Figs 2, 4, 5) but did not conform to any of these consensus phosphorylation motifs. Ser216 and Ser299 appear to be newly identified CDPK phosphorylation sites with an unknown CDPK phosphorylation motif. Whether Ser216, Thr296, and Ser299 are indeed phosphorylated by CDPK in vivo needs further confirmation.
The Arabidopsis genome has eight genes that encode active ACS proteins and an additional gene that encodes a catalytically inactive enzyme, ACS1 (Liang et al., 1992). Previous studies showed that ACS1 was inactive because it lacked the highly conserved tripeptide Thr-Asn-Pro (TNP) between Ile204 and Ser205 in the ACS conserved region that was responsible for binding the cofactor PLP (Liang et al., 1995; Yamagami et al., 2003). The introduction of TNP into ACS1 restores ACS activity, whereas its removal from enzymatically active ACS2 results in a loss of activity (Liang et al., 1995). In the sequence alignment of plant ACS proteins (data not shown), it was found that ACS7 has a tripeptide, Thr-Asn-Pro (TNP), between Ile212 and Ser216. Ser216 was phosphorylated by CDPK16 in the in vitro kinase assay (Figs 2, 4), and the phosphorylation of Ser216 in ACS7 can probably influence the interaction between ACS7 and its cofactor PLP and finally change the enzyme activity. To investigate whether phosphorylation can influence enzyme activity, an in vitro enzyme activity assay was performed according to previous studies (Li et al., 1996; Chae et al., 2003). The results indicated that non-phosphorylated ACS7 had a K m value of 27.43 μM, whereas phosphorylated ACS7 had a lower K m value (13.65 μM; data not shown). These results suggest that phosphorylated ACS7 has a higher catalytic ability, and the phosphorylation of ACS7 catalysed by CDPK16 may enhance ACS7 activity. However, such a possibility requires further studies.
ACS gene expression in light-grown mature Arabidopsis plants was previously studied using reverse transcription–PCR (RT–PCR) (Yamagami et al., 2003), in which the expression of all Arabidopsis ACS genes was surveyed in plant roots, leaves, stems, flowers, and siliques. These researchers found that ACS7 was expressed only in roots. It was confirmed here that ACS7 is a cytosolic protein (Fig. 6), suggesting that ACS7 protein is mainly localized in the root cytosol. Another study used a green fluorescent protein (GFP) fused to nine kinds of AtCDPKs (Dammann et al., 2003). The authors found that AtCDPK16 was localized to the root plasma membrane in a thin layer at the periphery of the cell. Further studies are needed to confirm whether ACS7 can be phosphorylated by CDPK16 in plant roots in vivo.
Lithium chloride is known to suppress root curvature in Pisum sativum seedlings (Belyavskaya, 2001). In Arabidopsis, the root cap comprises four tiers of columella cells that originate from initial columella cells. Columella cells have a specific polarity, in which the nucleus and ER are localized to the proximal side of the root meristem and periphery of the cell, respectively. In addition to the peripheral arrangement of the ER, its function as an intracellular calcium reservoir has been the preferred hypothesis for a possible gravity-sensing mechanism in columella cells, in which amyloplasts that come into contact with the peripheral ER can trigger the release of calcium stored in the ER (Perbal and Driss-Ecole, 2003). Many signalling molecules and secondary messengers, such as calcium (Plieth and Trewavas, 2002) and InsP3 (Perera et al., 2001), and pH (Johannes et al., 2001) have been implicated in linking gravity sensing to the initiation of a differential growth response. Very rapid gravity-specific changes in transcript abundance have been documented in gravistimulated root tips of Arabidopsis (Kimbrough et al., 2004) and whole seedlings (Moseyko et al., 2002). InsP3 is well known to induce calcium release from intracellular calcium reservoirs. Changes in InsP3 levels have been reported after the gravistimulation of pulvini in maize (Perera et al., 1999). Lithium chloride has been reported to be a phosphatidylinositol cycle inhibitor (Belyavskaya, 2001; Dieck et al., 2012). It can suppress InsP3 cycling and signalling by inhibiting inositol-1-phosphatase (Belyavskaya, 2001), leading to a reduction in the amount of InsP3. When plants are treated with LiCl, some plant organs, such as tonoplasts and the ER, that are reported to have IP3-sensitive calcium channels may be blocked, leading to a reduction of calcium ion efflux from these organs. When the gravity vector changes, InsP3 may bind to the ER or tonoplast InsP3-sensitive calcium channels, which ultimately leads to calcium ion efflux from these organs. These calcium ions are captured by some calcium sensors, such as CDPK and CaM, and pass the signals to downstream effectors, ultimately leading to the plant gravity response.
Several mutant lines have been shown to be related to calcium signalling in root gravitropism in plants. A mutation of the calmodulin agr-3 gene exerted a reduced root gravitropism phenotype in Arabidopsis (Sinclair et al., 1996). Spalding’s group found that a mutation of the Glutamate Receptor-Like 3.3 gene (i.e. a mammalian calcium channel orthologue) showed a root gravitropism phenotype (Miller et al., 2010). Moreover, an Arabidopsis calcium-dependent binding protein AtCLB gene overexpression line exhibited reduced root gravitropism, but a T-DNA insertion mutant exhibited greater root gravitropism than the WT (de Silva et al., 2011). However, little is known about whether the ACS gene is involved in calcium signalling. Additionally, these mutant lines expressed a root gravitropism phenotype without EGTA treatment. In contrast, the present study observed root gravitropism with EGTA treatment. Therefore, the root gravitropism phenotype observed here appeared to be distinct from others.
Auxin polar transport is involved in many developmental processes, such as vascular differentiation and tropic growth (Luschnig et al., 1998; Mattson et al., 1999; Rashotte et al., 2000). The auxin polar transport inhibitor NPA was used to investigate the role of the ACS7 gene in the gravity response using a T-DNA knockout acs7-1 mutant line and it was found that acs7-1 is more resistant to NPA-mediated inhibition (Fig. 11). The data indicate that the inhibition of the gravity response by NPA was greatly attenuated by the addition of 0.1 μM exogenous ACC (Fig. 11). Collectively, these findings suggest cross-talk between auxin and ethylene in root gravitropism regulated by AtACS7. Cross-talk between the phytohormone auxin and ethylene in root gravitropism has been reported previously. In 1990, Estelle’s group found that an aux1 mutant exhibited a defect in root gravitropism and resistance to both auxin and ethylene in Arabidopsis (Pickett et al., 1990). In 2003, an alh1 mutation revealed cross-talk between ethylene and auxin in Arabidopsis (Vandenbussche et al., 2003). However, how auxin engages in cross-talk with ethylene to regulate root gravitropism through AtACS7 in Arabidopsis requires further investigation.
The rice type II ACC synthase OsACS1 was found to interact with rice 14-3-3 proteins in a yeast two-hybrid assay (Yao et al., 2007). These authors found that the C-terminal domain of OsACS1 (RSVpSCP) was predicted to be phosphorylated by CDPK through a Mode-I 14-3-3 recognition motif. They suggested that OsACS1 phosphorylation catalysed by OsCDPK may interact with rice 14-3-3 proteins, which can prevent the substrate ubiquitin adaptor protein ETO1 from binding to produce 26S proteasome degradation. Interactions with 14-3-3 proteins on the CDPK phosphorylation site may prevent ETO1 protein binding, and this may be a reason why the phosphorylation of ACS proteins catalysed by CDPK enhances ACS protein stability. In fact, the phosphorylation status of AtACS6 affected the stability of AtACS6 (Skottke et al., 2011). Moreover, the stability of AtACS5 was also found to be regulated by phosphorylation and 14-3-3 binding (Yoon and Kieber, 2013). A recent study found that ACS7 is turned over in a 26S proteasome-dependent manner and that ACS7 degradation requires the E3 ligase XBAT32 (Lyzenga et al., 2012). Whether phosphorylation and 14-3-3 binding are involved in the regulation of AtACS7 degradation requires further investigation.
Apart from ACS, ACO is another important enzyme in the ethylene biosynthesis pathway that catalyses the oxidation of ACC to ethylene. ACO is encoded by a small multigene family that is usually composed of three to four members. In the Arabidopsis genome, six genes that encode ACOs were identified in silico by Babula et al. (2006). Recently, ACO2 and ACO4 proteins were found, which may interact with 14-3-3 proteins in a yeast two-hybrid system (Jaspert et al., 2011). Together with the present results, this indicates that 14-3-3 proteins may regulate the ethylene biosynthesis pathway by modulating ACS and ACO proteins. A previous study showed that an ACO promoter can respond to IAA, bending stress, and gravity (Yuan and Dean, 2010). Together, ACS proteins and ACO may synergistically participate in the gravity response by regulating ethylene concentrations in plant cells.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Plasmid maps used in the present study.
Figure S2. Gravity response measurement method.
Figure S3. Bar graph of the phosphorylation signal in the in vitro kinase assay using mutated AtCDPK16.
Figure S4. MS/MS fragmentation pattern of a phosphorylated peptide (VGTIYSYNDNVVR) of ACS7 recombinant protein.
Figure S5. Protein–protein interaction between ACS7 and 14-3-3ω using BiFC analysis.
Figure S6. Quartz crystal microbalance (QCM) and kinetic analysis of protein–protein interaction between ACS7 and 14-3-3ω recombinant proteins.
Figure S7. Isolation of the T-DNA insertional mutant acs7-1.
Figure S8. Gravity response of the WT and acs7-1 mutant affected by 0.5mM LaCl3.
Figure S9. The effect of 50 μM CPZ on the WT and acs7-1 mutant.
Table S1. Information on fusion peptides.
Acknowledgements
We thank Keqing Wu (National Taiwan University, Taiwan) for providing the pEarleyGate201-YN and pEarleyGate202-YC vectors. We thank Jeffrey Harper (University of Nevada, Reno, NV, USA) for providing GST-CDPK-6His plasmids. We appreciate the funding support from National Taiwan University (101R892002 and 102R892002) and the National Science Council, Taiwan (NSC#101-2321-B-002-049 and NSC#102-2311-B-002-029) to I-FC. We also thank Technology Commons, College of Life Science, National Taiwan University, and Technology Commons of Life Science and Precision Instrumentation Center sponsored by National Science Council, National Taiwan University for technical support. The English has been edited and proofread by BioMed Proofreading, LLC.
References
- Adams DO, Yang SF. 1979. Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proceedings of the National Academy of Sciences, USA 76, 170–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babula D, Misztal LH, Jakubowicz M, Kaczmarek M, Nowak W, Sadowski J. 2006. Genes involved in biosynthesis and signalisation of ethylene in Brassica oleracea and Arabidopsis thaliana: identification and genome comparative mapping of specific gene homologues. Theoretical and Applied Genetics 112, 410–420 [DOI] [PubMed] [Google Scholar]
- Batistic O, Kudla J. 2012. Analysis of calcium signaling pathways in plants. Biochimica et Biophysica Acta 1820, 1283–1293 [DOI] [PubMed] [Google Scholar]
- Belyavskaya NA. 2001. Lithium-induced changes in gravicurvature, statocyte ultrastructure and calcium balance of pea roots. Advances in Space Research 27, 961–966 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Peer WA, Makam SN, Murphy AS. 2004. Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiology 134, 28–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44 [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry 72, 248–254 [DOI] [PubMed] [Google Scholar]
- Buer CS, Sukumar P, Muday GK. 2006. Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiology 140, 1384–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HS, Faure F, Kieber JJ. 2003. The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. The Plant Cell 15, 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HS, Kieber JJ. 2005. Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends in Plant Science 10, 291–296 [DOI] [PubMed] [Google Scholar]
- Chang IF, Curran A, Woolsey R, Quilici D, Cushman JC, Mittler R, Harmon A, Harper JF. 2009. Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana . Proteomics 9, 2967–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Hilson P, Sedbrook J, Rosen E, Caspar T, Masson PH. 1998. The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proceedings of the National Academy of Sciences, USA 95, 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker W, Knight LL. 1908. Effect of illuminating gas and ethylene upon flowering carnations. Botanical Gazette 46, 259–276 [Google Scholar]
- Curran A, Chang IF, Chang CL, et al. 2011. Calcium-dependent protein kinases from Arabidopsis show substrate specificity differences in an analysis of 103 substrates. Frontiers in Plant Science 2, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann C, Ichida A, Hong B, Romanowsky SM, Hrabak EM, Harmon AC, Pickard BG, Harper JF. 2003. Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiology 132, 1840–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depta H, Rubery PH. 1984. A comparative study of carrier participation in the transport of 2,3,5-triiodobenzoic acid, indole-3-acetic-acid, and 2,4-dichlorophenoxyacetic acid by Cucurbita pepo L. hypocotyl segments. Journal of Plant Physiology 115, 371–387 [DOI] [PubMed] [Google Scholar]
- de Silva K, Laska B, Brown C, Sederoff HW, Khodakovskaya M. 2011. Arabidopsis thaliana calcium-dependent lipid-binding protein (AtCLB): a novel repressor of abiotic stress response. Journal of Experimental Botany 62, 2679–2689 [DOI] [PubMed] [Google Scholar]
- Dieck CB, Boss WF, Perera IY. 2012. A role for phosphoinositides in regulating plant nuclear functions. Frontiers in Plant Science 3, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D. 2010. The language of calcium signaling. Annual Review of Plant Biology 61, 593–620 [DOI] [PubMed] [Google Scholar]
- Dong H, Zhen ZQ, Peng JY, Chang L, Gong QQ, Wang NN. 2011. Loss of ACS7 confers abiotic stress tolerance by modulating ABA sensitivity and accumulation in Arabidopsis. Journal of Experimental Botany 62, 4875–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano JM, Swanson SJ, Blancaflor EB, Dowd PE, Kao TH, Gilroy S. 2001. Changes in root cap pH are required for the gravity response of the Arabidopsis root. The Plant Cell 13, 907–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Grosskopf DG, Regenass M, Basse CW, Boller T. 1991. Elicitor-induced ethylene biosynthesis in tomato cells—characterization and use as a bioassay for elicitor action. Plant Physiology 97, 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Piconese S, Tronelli G, Migliaccio F. 2000. A new Arabidopsis thaliana root gravitropism and chirality mutant. Plant Science 125, 990–1000 [DOI] [PubMed] [Google Scholar]
- Fortunati A, Piconese S, Tassone P, Ferrari S, Migliaccio F. 2008. A new mutant of Arabidopsis disturbed in its roots, right-handed slanting, and gravitropism defines a gene that encodes a heat-shock factor. Journal Experimental Botany 59, 1363–1374 [DOI] [PubMed] [Google Scholar]
- Friedman H, Meir S, Rosenberger I, Halevy AH, Kaufman PB, Philosoph-Hadas S. 1998. Inhibition of the gravitropic response of snapdragon spikes by the calcium-channel blocker lanthanum chloride. Plant Physiology 118, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G. 2003. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426, 147–153 [DOI] [PubMed] [Google Scholar]
- Grosskopf DG, Felix G, Boller T. 1990. K-252a inhibits the response of tomato cells to fungal elicitors in vivo and their microsomal protein kinase in vitro . FEBS Letters 275, 177–180 [DOI] [PubMed] [Google Scholar]
- Guzman P, Ecker JR. 1990. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. The Plant Cell 2, 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Harmon A. 2005. Plants, symbiosis and parasites: a calcium signaling connection. Nature Reviews Molecular Cell Biology 6, 555–566 [DOI] [PubMed] [Google Scholar]
- Hasenstein KH, Evans ML. 1988. Effects of cations on hormone transport in primary roots of Zea mays . Plant Physiology 86, 890–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegeman AD, Rodriguez M, Han BW, Uno Y, Phillips GN, Jr, Hrabak EM, Cushman JC, Harper JF, Harmon AC, Sussman MR. 2006. A phyloproteomic characterization of in vitro autophosphorylation in calcium-dependent protein kinases. Proteomics 6, 3649–3664 [DOI] [PubMed] [Google Scholar]
- Hetherington AM, Trewavas A. 1984. Activation of a pea membrane protein kinase by calcium ions. Planta 161, 409–417 [DOI] [PubMed] [Google Scholar]
- Hoson T, Kamisaka S, Masuda Y. 1996. Suppression of gravitropic response of primary roots by submergence. Planta 199, 100–104 [DOI] [PubMed] [Google Scholar]
- Huang JZ, Huber SC. 2001. Phosphorylation of synthetic peptides by a CDPK and plant SNF1-related protein kinase. Influence of proline and basic amino acid residues at selected positions. Plant and Cell Physiology 42, 1079–1087 [DOI] [PubMed] [Google Scholar]
- Jaspert N, Throm C, Oecking C. 2011. Arabidopsis 14-3-3 proteins: fascinating and less fascinating aspects. Frontiers of Plant Science 2, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes E, Collings DA, Rink JC, Allen NS. 2001. Cytoplasmic pH dynamics in maize pulvinal cells induced by gravity vector changes. Plant Physiology 127, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyoshihara Y, Iwata M, Fukaya T, Tatsuki M, Mori H. 2010. Turnover of LeACS2, a wound-inducible 1-aminocyclopropane-1-carboxylic acid synthase in tomato, is regulated by phosphorylation/dephosphorylation. The Plant Journal 64, 140–150 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. 1993. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441 [DOI] [PubMed] [Google Scholar]
- Kimbrough JM, Salinas-Mondragon R, Boss WF, Brown CS, Sederoff HW. 2004. The fast and transient transcriptional network of gravity and mechanical stimulation in the Arabidopsis root apex. Plant Physiology 136, 2790–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli GK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- Lee LY, Wu FH, Hsu CT, et al. 2012. Screening a cDNA library for protein–protein interactions directly in planta. The Plant Cell 24, 1746–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Huxtable S, Yang SF, Kung SD. 1996. Effects of N-terminal deletions on 1-aminocyclopropane-1-carboxylate synthase activity. FEBS Letters 378, 286–290 [DOI] [PubMed] [Google Scholar]
- Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S. 2012. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genetics 8, e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XW, Abel S, Keller JA, Shen NF, Theologis A. 1992. The 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 89, 11046–11050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Oono Y, Shen NF, Köhler C, Li K, Scolnik PA, Theologis A. 1995. Characterization of two members (ACS1 and ACS3) of the 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Gene 167, 17–24 [DOI] [PubMed] [Google Scholar]
- Liese A, Romeis T. 2012. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochimica et Biophysica Acta 1833, 1582–1589 [DOI] [PubMed] [Google Scholar]
- Lin ZF, Zhong SL, Grierson D. 2009. Recent advances in ethylene research. Journal of Experimental Botany 60, 3311–3336 [DOI] [PubMed] [Google Scholar]
- Liu YD, Zhang SQ. 2004. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. The Plant Cell 16, 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig AA, Saitoh H, Felix G, Freymark G, Miersch O, Wasternack C, Boller T, Jones JDG, Romeis T. 2005. Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proceedings of the National Academy of Sciences, USA 102, 10736–10741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. 1998. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana . Genes and Development 12, 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga WJ, Booth JK, Stone SL. 2012. The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. The Plant Journal 71, 23–34 [DOI] [PubMed] [Google Scholar]
- Madlung A, Behringer FJ, Lomax TL. 1999. Ethylene plays multiple nonprimary roles in modulating the gravitropic response in tomato. Plant Physiology 120, 897–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga H, Ueda H. 2010. Stress-induced non-vesicular release of prothymosin-α initiated by an interaction with S100A13, and its blockade by caspase-3 cleavage. Cell Death and Differentiation 17, 1760–1772 [DOI] [PubMed] [Google Scholar]
- Mattsson J, Sung ZR, Berleth T. 1999. Responses of plant vascular systems to auxin transport inhibition. Development 126, 2979–2991 [DOI] [PubMed] [Google Scholar]
- McClellan CA, Chang C. 2008. The role of protein turnover in ethylene biosynthesis and response. Plant Science 175, 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller ND, Durham Brooks TL, Assadi AH, Spalding EP. 2010. Detection of a gravitropism phenotype in glutamate receptor-like 3.3 mutants of Arabidopsis thaliana using machine vision and computation. Genetics 186, 585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita MT. 2010. Directional gravity sensing in gravitropism. Annual Review of Plant Biology 61, 705–720 [DOI] [PubMed] [Google Scholar]
- Moseyko N, Zhu T, Chang HS, Wang X, Feldman LJ. 2002. Transcription profiling of the early gravitropic response in Arabidopsis using high-density oligonucleotide probe microarrays. Plant Physiology 130, 720–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva M, Dunand C. 2007. Waving and skewing: how gravity and the surface of growth media affect root development in Arabidopsis. New Phytologist 176, 37–43 [DOI] [PubMed] [Google Scholar]
- Peng HP, Lin TY, Wang NN, Shih MC. 2005. Differential expression of genes encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis during hypoxia. Plant Molecular Biology 58, 15–25 [DOI] [PubMed] [Google Scholar]
- Perbal G, Driss-Ecole D. 2003. Mechanotransduction in gravisensing cells. Trends in Plant Science 8, 498–504 [DOI] [PubMed] [Google Scholar]
- Perera IY, Heilmann I, Boss WF. 1999. Transient and sustained increases in inositol 1,4,5-trisphosphate precede the differential growth response in gravistimulated maize pulvini. Proceedings of the National Academy of Sciences, USA 96, 5838–5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Heilmann I, Chang SC, Boss WF, Kaufman PB. 2001. A role for inositol 1,4,5-trisphosphate in gravitropic signaling and the retention of cold-perceived gravistimulation of oat shoot pulvini. Plant Physiology 125, 1499–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Hung CY, Brady S, Muday GK, Boss WF. 2006. A universal role for inositol 1,4,5-trisphosphate-mediated signaling in plant gravitropism. Plant Physiology 140, 746–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perochon A, Aldon D, Galaud JP, Ranty B. 2011. Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie 93, 2048–2053 [DOI] [PubMed] [Google Scholar]
- Philosoph-Hadas S, Meri S, Meir S, Rosenberger I, Halevy AH. 1996. Regulation of the gravitropic response and ethylene biosynthesis in gravistimulated snapdragon spikes by calcium chelators and ethylene inhibitors. Plant Physiology 110, 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett FB, Wilson AK, Estelle M. 1990. The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiology 94, 1462–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plieth C, Trewavas AJ. 2002. Reorientation of seedlings in the earth’s gravitational field induces cytosolic calcium transients. Plant Physiology 129, 786–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Brady SR, Reed RC, Ante SJ, Muday GK. 2000. Basipetal auxin transport is required for gravitropism in roots of Arabidopsis. Plant Physiology 122, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V, Fluhr R. 1992. Calcium requirement for ethylene-dependent responses. The Plant Cell 4, 1123–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Brownlee C, Harper JF. 1999. Communicating with calcium. The Plant Cell 11, 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF. 2002. Calcium at the crossroads of signaling. The Plant Cell 14, Suppl, S401–S417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisree P, Nongmaithem S, Vasuki H, Sreelakshmi Y, Ivanchenko MG, Sharma R. 2011. Tomato root penetration in soil requires a coaction between ethylene and auxin signaling. Plant Physiology 156, 1424–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastià CH, Hardin SC, Clouse SD, Kieber JJ, Huber SC. 2004. Identification of a new motif for CDPK phosphorylation in vitro that suggests ACC synthase may be a CDPK substrate. Archives of Biochemistry and Biophysics 428, 81–91 [DOI] [PubMed] [Google Scholar]
- Sedbrook J, Boonsirichai K, Chen R, et al. 1998. Molecular genetics of root gravitropism and waving in Arabidopsis thaliana . Gravitational and Space Biology Bulletin 11, 71–78 [PubMed] [Google Scholar]
- Sehnke PC, DeLille JM, Ferl RJ. 2002. Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. The Plant Cell 14, S339–S354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair W, Oliver I, Maher P, Trewavas A. 1996. The role of calmodulin in the gravitropic response of the Arabidopsis thaliana agr-3 mutant. Planta 199, 343–351 [DOI] [PubMed] [Google Scholar]
- Skottke KR, Yoon GM, Kieber JJ, DeLong A. 2011. Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genetics 7, e1001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu P, Grosskopf DG, Felix G, Boller T. 1994. The apparent turnover of 1-aminocyclopropane-1-carboxylate synthase in tomato cells is regulated by protein phosphorylation and dephosphorylation. Plant Physiology 106, 529–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohm AK, Baldwin KL, Masson PH. 2012. Molecular mechanisms of root gravity sensing and signal transduction. WIREs Developmental Biology 1, 276–285 [DOI] [PubMed] [Google Scholar]
- Sukumar P, Edwards KS, Rahman A, Delong A, Muday GK. 2009. PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiology 150, 722–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Dhonukshe P, Brewer PB, Friml J. 2006. Spatiotemporal asymmetric auxin distribution: a means to coordinate plant development. Cellular and Molecular Life Sciences 63, 2738–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuki M, Mori H. 2001. Phosphorylation of tomato 1-aminocyclopropane-1-carboxylic acid synthase, LE-ACS2, at the C-terminal region. Journal of Biological Chemistry 276, 28051–28057 [DOI] [PubMed] [Google Scholar]
- Thomson KS, Hertel R, Muller S, Tavares JE. 1973. 1-N-naphthylphthalamic acid and 2,3,5-triiodobenzoic acid—in vitro binding to particulate cell fractions and action on auxin transport in corn coleoptiles. Planta 109, 337–352 [DOI] [PubMed] [Google Scholar]
- Toyota M, Furuichi T, Tatsumi H, Sokabe M. 2008. Critical consideration on the relationship between auxin transport and calcium transients in gravity perception of Arabidopsis seedlings. Plant Signaling and Behavior 3, 521–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas A. 1999. Le calcium, C’est la vie: calcium makes waves. Plant Physiology 120, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A, Theologis A. 2004. Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proceedings of the National Academy of Sciences, USA 101, 2275–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A, Yu GX, Jin HL, Alonso JM, Ecker JR, Zhang XM, Gao S, Theologis A. 2009. A combinatorial interplay among the 1-aminocyclopropane-1-carboxylate isoforms regulates ethylene biosynthesis in Arabidopsis thaliana . Genetics 183, 979–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsuno K, Shikanai T, Yamada Y, Hashimoto T. 1998. Agr, an Agravitropic locus of Arabidopsis thaliana, encodes a novel membrane-protein family member. Plant and Cell Physiology 39, 1111–1118 [DOI] [PubMed] [Google Scholar]
- Vandenbussche F, Smalle J, Le J, et al. 2003. The Arabidopsis mutant alh1 illustrates a cross talk between ethylene and auxin. Plant Physiology 131, 1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Li H, Ecker JR. 2002. Ethylene biosynthesis and signaling networks. The Plant Cell 14, Supplement, S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Yoshida H, Lurin C, Ecker JR. 2004. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428, 945–950 [DOI] [PubMed] [Google Scholar]
- Went FA. 1928. The international union of biological sciences. Science 68, 545–547 [DOI] [PubMed] [Google Scholar]
- Wolverton C, Paya AM, Toska J. 2011. Root cap angle and gravitropic response rate are uncoupled in the Arabidopsis pgm-1 mutant. Physiologia Plantarum 141, 373–382 [DOI] [PubMed] [Google Scholar]
- Wu K, Lu GH, Sehnke P, Ferl RJ. 1997. The heterologous interactions among plant 14-3-3 proteins and identification of regions that are important for dimerization. Archives of Biochemistry and Biophysics 339, 2–8 [DOI] [PubMed] [Google Scholar]
- Yamagami T, Tsuchisaka A, Yamada K, Haddon WF, Harden LA, Theologis A. 2003. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. Journal of Biological Chemistry 278, 49102–49112 [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. 1984. Ethylene biosynthesis and its regulation in higher plants. Annual Review of Plant Physiology 35, 155–189 [Google Scholar]
- Yao Y, Du Y, Jiang L, Liu JY. 2007. Interaction between ACC synthase 1 and 14-3-3 proteins in rice: a new insight. Biochemistry (Moscow) 72, 1003–1007 [DOI] [PubMed] [Google Scholar]
- Yip WK, Dong JG, Yang SF. 1991. Purification and characterization of 1-aminocyclopropane-1-carboxylate synthase from apple fruits. Plant Physiology 95, 251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho Y, Sheen J. 2009. Emerging connections in the ethylene signaling network. Trends in Plant Science 14, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon GM, Kieber JJ. 2013. 14-3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. The Plant Cell 25, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Nagata M, Saito K, Wang KL, Ecker JR. 2005. Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biology 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Wang LC, Chang CM, Mori K, Uchida E, Ecker JR. 2006. The ACC synthase TOE sequence is required for interaction with ETO1 family proteins and destabilization of target proteins. Plant Molecular Biology 62, 427–437 [DOI] [PubMed] [Google Scholar]
- Yuan S, Dean JF. 2010. Differential responses of the promoters from nearly identical paralogs of loblolly pine (Pinus taeda L.) ACC oxidase to biotic and abiotic stresses in transgenic Arabidopsis thaliana . Planta 232, 873–886 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.