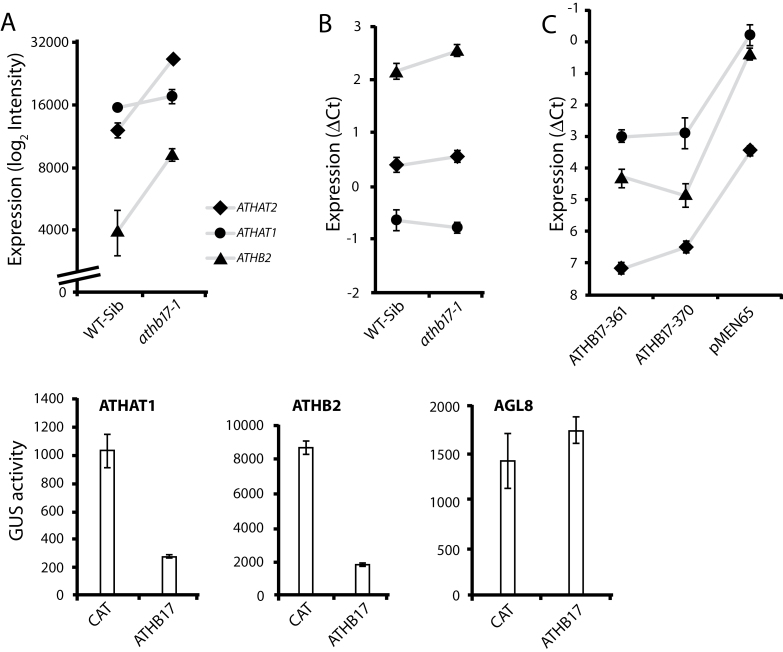
Fig. 2.
Expression levels of ATHAT2, ATHAT1, and ATHB2 in various Arabidopsis tissues. Microarray analysis indicated that the transcript levels of selected class II HB-Zip genes were altered in the QC-enriched samples of athb17-1 compared with its wild-type sibling. (A) ATHAT2 and ATHB2 expression, but not ATHAT1 expression, was induced in the athb17-1 QC. (B) qRT-PCR was used to verify the altered expression pattern observed in panel A. (C) ATHAT1, ATHAT2, and ATHB2 expression is significantly repressed in seedlings of two 35S::ATHB17 lines (line 361 and line 370). (D) ATHB17 also repressed the transcription of ATHAT1 and ATHB2 in Arabidopsis mesophyll protoplasts. Full-length ATHB17 or CAT was co-transfected with the pATHAT1::GUS or pATHB2::GUS reporter gene into Arabidopsis mesophyll protoplasts. The pAGL8::GUS reporter gene was used as a non-specific reporter as it was not expected to be regulated by ATHB17. The data are means ±standard deviation (SD) of three replicates. PCR comparisons were made after normalization to a constitutive control gene by the ΔCt method.