Figure 3.
The Proteolytic Site of CtpB Is Sequestered in a Narrow Tunnel
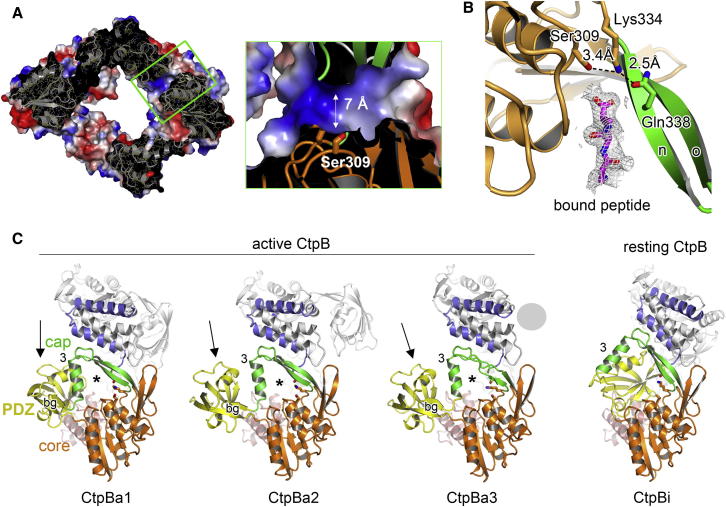
(A) Surface presentation of the CtpB dimer with mapped electrostatic potential. The molecule is shown in a half-cut view displaying the construction of one of the two protease tunnels penetrating the dimer. The zoomed-in window illustrates the position of the catalytic serine buried within the tunnel, as well as the dimensions of the molecular passage.
(B) The catalytic residues Ser309, Lys334, and Gln338 are shown in stick representation and their respective distances are indicated. The omit electron density of a cocrystallized ligand (1.9 Å resolution, contoured at 1.2 σ) is shown together with a modeled poly-Ala peptide.
(C) Ribbon plots of CtpBa1, CtpBa2, CtpBa3, and CtpBi. To illustrate the pronounced rearrangements of the protease cap, helix α3 is labeled. Moreover, the variable positions of the PDZ domain are indicated by an arrow, the opening of the proteolytic tunnel is marked with an asterisk and the β sheet b-g, which was only observed in the active conformations, is labeled. In CtpBa3, one PDZ domain was too flexible to be modeled into electron density and is indicated by a gray sphere. See also Figure S3.