Figure S5.
Peptide Binding to the PDZ Domain Stabilizes the Active Protease State of CtpB, Related to Figure 5
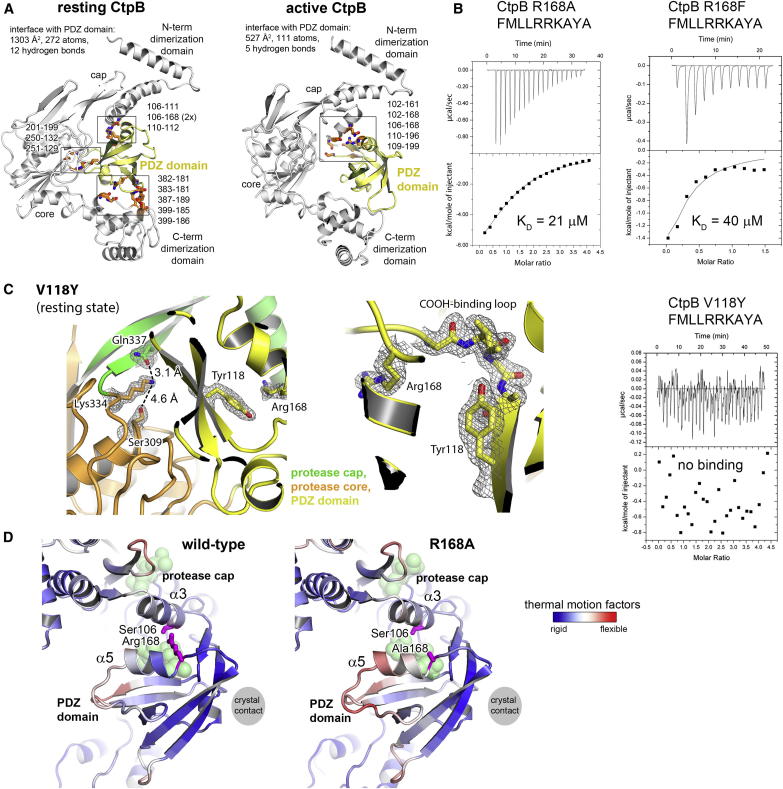
(A) The interfaces of the PDZ domain with the other domains of CtpB were analyzed to evaluate the prevailing conformation of CtpB in solution. The ribbon representations of CtpBi (left) and CtpBa1 (right) emphasize the PDZ domain that is shown in yellow. Interactions stabilizing the respective domain interfaces (summarized in top panel) are highlighted in orange and the residues forming hydrogen-bonds are listed. Interactions of the PDZ domain with protease core and C-terminal dimerization domain are only present in the resting state yielding a markedly increased domain interface. Accordingly, the resting state should be the prevailing conformation in solution, as also suggested by the protease assays. (B) ITC analysis characterizing the interaction of a peptide having an Ala-Tyr-Ala C terminus with the full-length CtpB variants R168A, R168F and V118Y. Raw data (top) and binding isotherm derived from the integrated heat (bottom) are shown. The determined KD dissociation constants are indicated below the binding curves. (C) Left: a ribbon model of the CtpB V118Y mutant structure, which was crystallized in the same resting conformation as CtpBi. Omit electron densities (1.95 Å resolution, contoured at 1.2 σ) of the catalytic triad and of the introduced Tyr118 are shown together with the respective stick models. Interatomic distances among the Ser309-Lys334-Gln338 active site residues are given and reveal that the OH group of Ser309 is located too far away (4.6 Å) to setup a functional catalytic triad. Accordingly, the V118Y crystal structure supports the model that peptide binding to the PDZ domain is critical to remodel CtpB into its active form, whereas the composition of the proteolytic site does not define the functional states. Middle: a zoomed-in image of Tyr118 that occupies a large part of the peptide-binding cleft of the PDZ domain protruding toward the indicated carboxylate-binding loop. Corresponding omit electron densities are shown. (D) Comparison of the thermal motion factors of the PDZ domain of wild-type CtpB and R168A mutant. Both proteins crystallized in the same crystal form thus experiencing a similar crystal contact, indicated by the gray sphere.