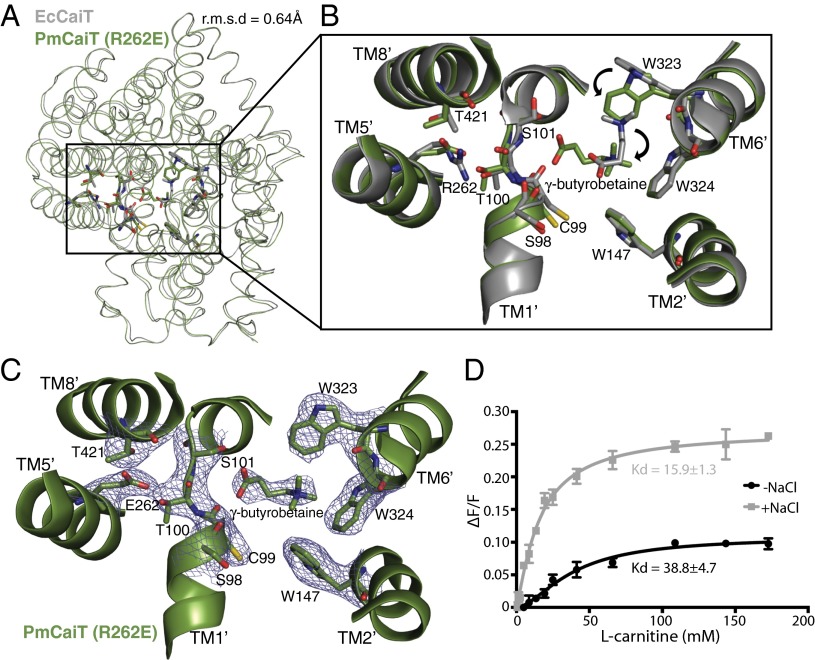
Fig. 3.
Substrate binding in CaiT R262 mutants. (A) Superimposition of EcCaiT (chain A) and PmCaiT R262E (PDB codes 2WSW and 4M8J, respectively) reveals an rmsd of 0.64 Å. (B) Central substrate binding site in the PmCaiT R262E structure with a bound γ-butyrobetaine superimposed on the substrate-bound EcCaiT type structure. In the R262E mutant, the substrate has rotated by about 90° compared with the wild-type structure. In addition, W323 has reoriented into a position suitable to accommodate the substrate in this new orientation. (C) 2Fo-Fc map of the central binding site at 1.0 σ. The trimethyl ammonium head group of the substrate is coordinated by the tryptophan box from TM6′ and TM2′, whereas the carboxyl group of the substrate is interacting with the backbone carbonyl of S98 in the unwound TM1′ helix. The glutamate at position 262 establishes a hydrogen bond to the unwound TM1′ helix, which, together with T421 from TM8′, stabilizes the unwound stretch, similar to Arg262 in the wild-type structure. (D) Tryptophan fluorescence-based substrate-binding assay with R262E reconstituted into liposomes. The increase in tryptophan fluorescence with rising l-carnitine concentration was monitored in the presence and absence of 50 mM NaCl. The relative increase in fluorescence was plotted using a one-site binding model to obtain the apparent Kd values for substrate binding. All data points are the averages of three independent experiments; the error bars indicate the SD of the measurements.