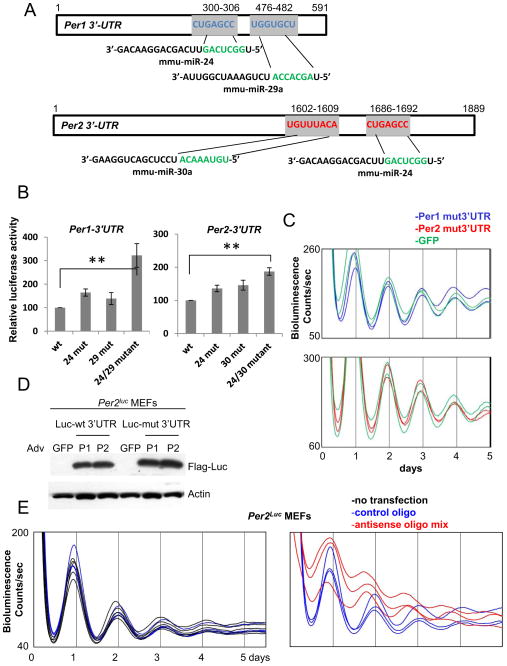
Fig 6.
miR-24/29a and 24/30a post-transcriptionally regulate Per1 and Per2, respectively. (A) Per1 and Per2 3′-UTRs have conserved target sites for miR-24/29a and 24/30a, respectively. The target sites were identified by TargetScanMouse 5.2. Note that these target sites have perfect 7 or 8 nucleotide matches. See also Fig S4. (B) Luciferase expression is higher when the target sites are mutated in transient transfection-based reporter assays. The mutant 3′-UTRs in the pGL3 basic plasmid were expressed in NIH3T3 cells and their expression levels were compared. Single mutations only modestly increased luciferase expression, but double mutations increased the luciferase expression significantly higher compared to wt UTRs. See also Fig S5A. (C) Overexpression of mutant miR-24/29a Per1 3′-UTR or mutant miR-24/30a Per2 3′-UTR does not induce period shortening. The mutations in the miRNA target sites were introduced into the adenoviral 3′-UTR constructs used in Fig 4. (D) Expression levels of these mutant UTRs were comparable to those of wt UTRs when expression levels of the partial luciferase attached to the UTRs were compared. (E) Circadian period is shortened in wt Per2Luc cells treated with antisense oligonucleotides to miR-24, 29a and 30a. A negative control oligonucleotide did not affect circadian period compared to non-treated control cells (left panel). The periods of bioluminescence rhythms in control and specific oligonucleotide-treated cells were 24.25+/−0.08 and 22.88+/−0.10, respectively (mean+/−SEM of 8 traces each; p<0.001). See also Fig S5B.