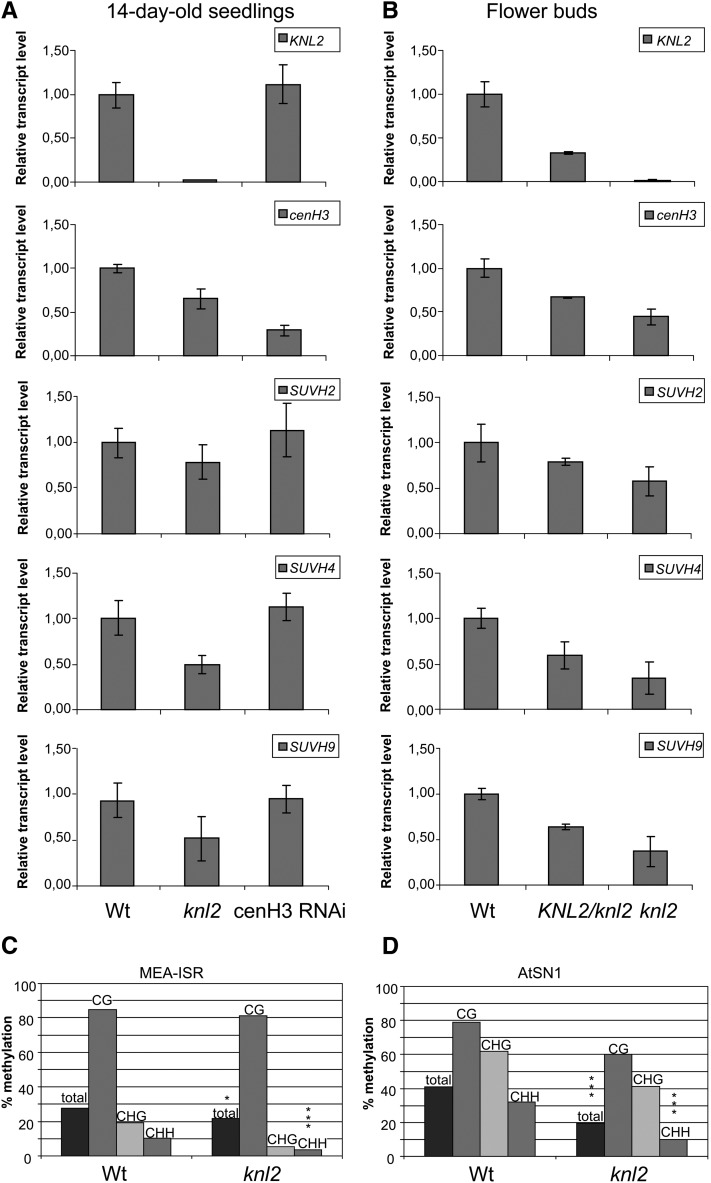
Figure 6.
Expression of KNL2, cenH3, SUVH2, SUVH4, and SUVH9 in the knl2 Mutant and cenH3 RNAi Plants and DNA Methylation at MEA-ISR and At-SN1 Loci in knl2 Mutants.
(A) and (B) Real-time quantitative RT-PCR analysis of KNL2, cenH3, SUVH2, SUVH4, and SUVH9 mRNA transcripts in 14-d-old seedlings of the knl2 mutant and cenH3 RNAi transformants compared with the wild type (A) and in flower buds of heterozygous KNL2/knl2 and homozygous knl2 mutants compared with the wild type (B). Amplification of ACTIN2 mRNA was used for data normalization.
(C) and (D) DNA methylation was determined by bisulfite sequencing of genomic DNA of 7-d-old seedlings. Individual clones were sequenced for the different genotypes. The bars mark the levels of DNA methylation in percentage of methylated cytosines relative to total cytosines. Wt, wild-type plants (Columbia-0).
(C) DNA methylation at MEA-ISR. Significant differences in total DNA methylation between the different genotypes were checked by χ2 tests. Wild type, n = 26; knl2, n = 19 (complete methylation: χ2, 5.54, *P < 0.05; asymmetric methylation: χ2, 18.24, ***P < 0.001).
(D) DNA methylation at At-SN1. Wild type, n = 12; At-knl2, n = 10 (χ2, 45.48, ***P < 0.001; asymmetric methylation: χ2, 51.30, ***P < 0.001).