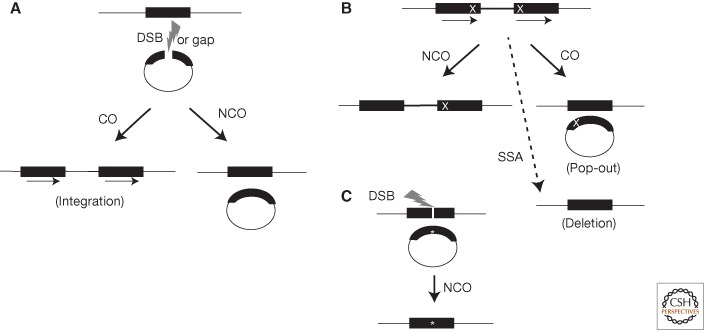
Figure 1.
HR events. (A) Plasmid-by-chromosome HR initiated by a DSB or gap (lightning bolt) in the plasmid. The black bars represent the homology between the chromosome and plasmid. In this case, the chromosomal sequence is used as the donor template to repair the gap in the recipient plasmid. Crossovers (COs) lead to plasmid integration and the formation of direct repeats (indicated by arrows). Noncrossovers (NCOs) are also detected if the plasmid contains an origin of replication. (B) Recombination between direct repeats is frequently used to assay HR. Different mutations (X) are present in each repeat. HR associated with gene conversion of one mutation leads to restoration of one intact repeat (solid black bars), which is wild type. NCOs maintain the direct repeat configuration, whereas a CO leads to a plasmid “pop-out” event. Whereas bona fide HR events (i.e., those involving gene conversion) maintain both repeats, SSA (for details, see Fig. 2) leads to deletion of one repeat and the segment between the repeats. (C) DSB-induced gene targeting. Because nonhomologous integration of plasmids in mammalian cells is so efficient, creation of a site-specific DSB in the chromosome is used to induce HR. For the purposes of genome modification (*), the repair of this DSB can be from an incoming plasmid donor sequence, which can be circular (shown) or linear (not shown).