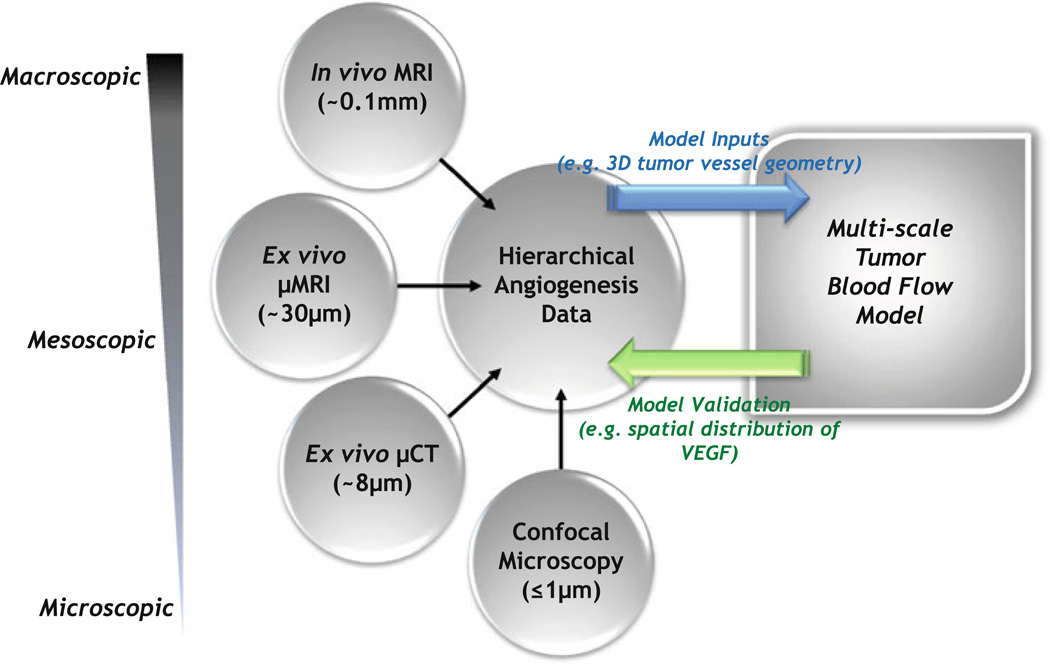
FIGURE 3.
Schematic illustrating the bidirectional feedback between multiscale imaging and multiscale modeling of tumor blood flow. Following acquisition of the hierarchical angiogenesis data, certain imaging parameters will serve as inputs to the multiscale model of tumor blood flow, while others will be employed to validate model predictions. For example, the µCT-derived 3-D tumor vessel geometry could serve as the input for large-scale tumor blood flow simulations. Predictions of the blood flow simulations will inform an oxygen transport model, which in turn will determine results of a VEGF secretion model. The VEGF secretion model will predict the spatio-temporal distribution of VEGF within the tumor, which will be validated against the spatial distribution of VEGF obtained from confocal microscopy of immunostained tumor sections. It should be borne in mind that acquiring macroscopic 2D (1 mm thick slices) in vivo MRI data over a 1–2 cm axial FOV at 125 µm in-plane resolution takes 5–10 min and results in ~MB of data. In contrast, a mesoscopic µMRI acquisition of the same volume at ~40–50 µm isotropic resolution takes 8–10 h, resulting in ~100 MB of data, while a microscopic µCT acquisition of the same volume at ~8 µm isotropic resolution takes 10–12 h, resulting in ~1 GB of data.