Fig. 6.
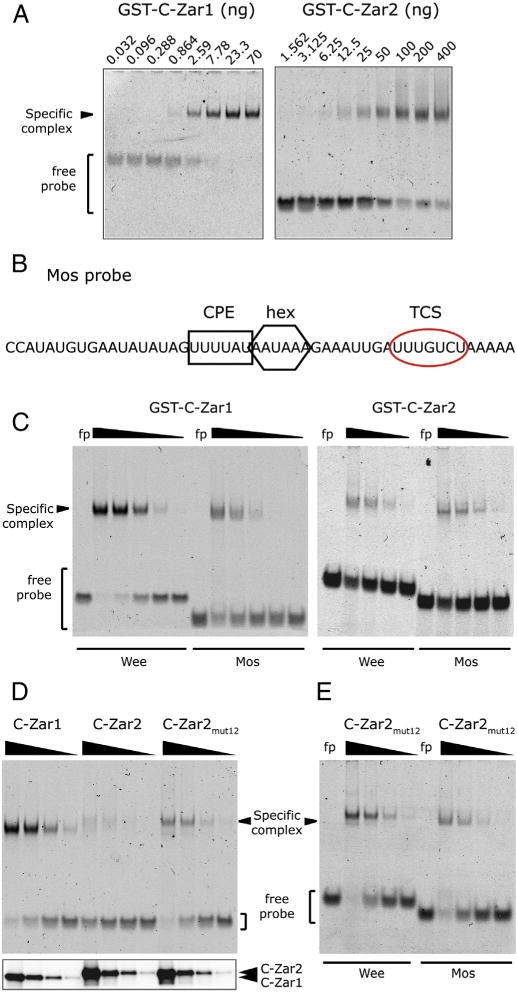
Comparison of RNA binding between Zar1 and Zar2. (A) Probe retardation in response to increasing amounts of bacterially expressed and purified Zar proteins. Left panel, GST-C-Zar1 was added to the binding reactions from 70 ng to 32 pg in a 3-fold dilution series. Right panel, GST-C-Zar2 was added to the binding reactions from 400 ng to 1.56 ng in a 2-fold dilution series. (B) Sequence of the Mos 3′ UTR probe used for this study. The CPE is marked with a black square, the polyadenylation hexanucleotide (hex) with a black hexagon, and the TCS with a red oval. (C) Differential binding of Zar1 and Zar2 to Wee and Mos 3′ UTRs. Left panel, Wee or Mos probes were incubated with decreasing amounts of GST-C-Zar1 from 100 ng in a 5-fold dilution series. Right panel, Wee or Mos probes were incubated with decreasing amounts of GST-C-Zar2 from 3 μg in a 5-fold dilution series. Zar1 binds more strongly to the Wee1 probe than to the Mos probe, while Zar2 binds to both probes with similar affinity. (D and E) Conserved amino acid differences between C-terminal domains of Zar1 and Zar2 contribute to RNA binding characteristics. (D) Upper panel, EMSA showing relative affinity of Zar proteins to the Wee1 UTR. 12 amino acids in GST-C-Zar2 were mutated to those in Zar1 as shown in Fig. 1 (C-Zar2mut12). Decreasing amounts of bacterially expressed GST-C-Zar1, GST-C-Zar2, or GST-C-Zar2mut12 from 625 ng in a 5-fold dilution series were incubated with Wee1 UTR. A representative gel of three different experiments is shown. Lower panel, western blot of the protein preparation, showing equivalent amounts of mutant proteins were used in the assay. (E) Differential binding of C-Zar2mut12 to Wee and Mos 3′ UTRs. Wee or Mos probes were incubated with decreasing amounts of GST-C-Zar2mut12 from 1250 ng in a 5-fold dilution series. Zar2mut12 binds a little more strongly to the Wee1 probe than to the Mos probe.
