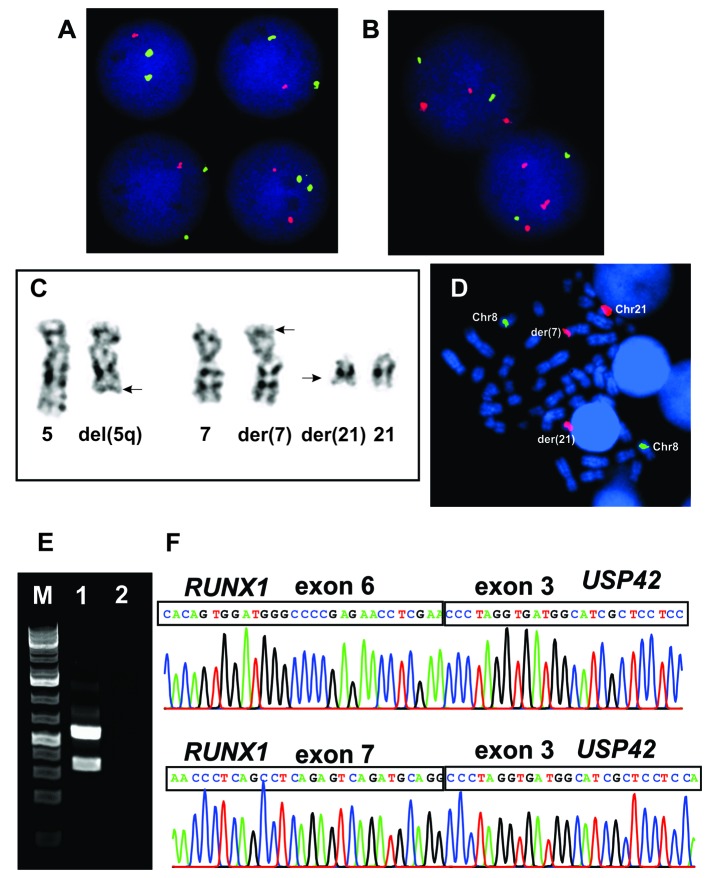
Figure 2.
Cytogenetic, FISH and PCR analyses. (A) Interphase FISH with del(5q) probe. The EGR1 probe (in 5q31) is labeled in red and the control probe mapped in 5p15.31 is labeled in green. Three nuclei had one red signal suggesting a hemizygous deletion of the EGR1 gene. All four nuclei had two green signals of the control probe. (B) Interphase FISH with the AML1/ETO probe. The AML1 probe (RUNX1) is labeled in red and the ETO probe (RUNX1T1) is labeled in green. Both nuclei had two green signals which suggest that the RUNX1T1 gene was not rearranged. Both nuclei had three red signals which suggest than one RUNX1 locus was rearranged. (C) Partial karyotype showing chromosome aberrations del(5q), der(7)t(7;21)(p22;q22), and der(21)t(7;21)(p22;q22) together with the corresponding normal homologues; breakpoint positions are indicated by arrows. (D) FISH on metaphase spread using the AML1/ETO probe. Green signals (ETO probe) are observed only on chromosomes 8 (normal RUNX1T1). Part of the AML1 probe (RUNX1) is located on 7p22. (E) cDNA fragment amplification of RUNX1-USP42 (lane 1) using the primers RUNX1–765F and USP42–562R. PCR with primers USP42–116F and RUNX1–1489R did not amplify any cDNA fragment (lane 2). M, 1 kb DNA ladder. (F) Partial sequence chromatograms of the two amplified RUNX1-USP42 fragments showing that exon 6 of RUNX1 is fused to exon 3 of USP42 and that exon 7 of RUNX1 is fused to exon 3 of USP42.