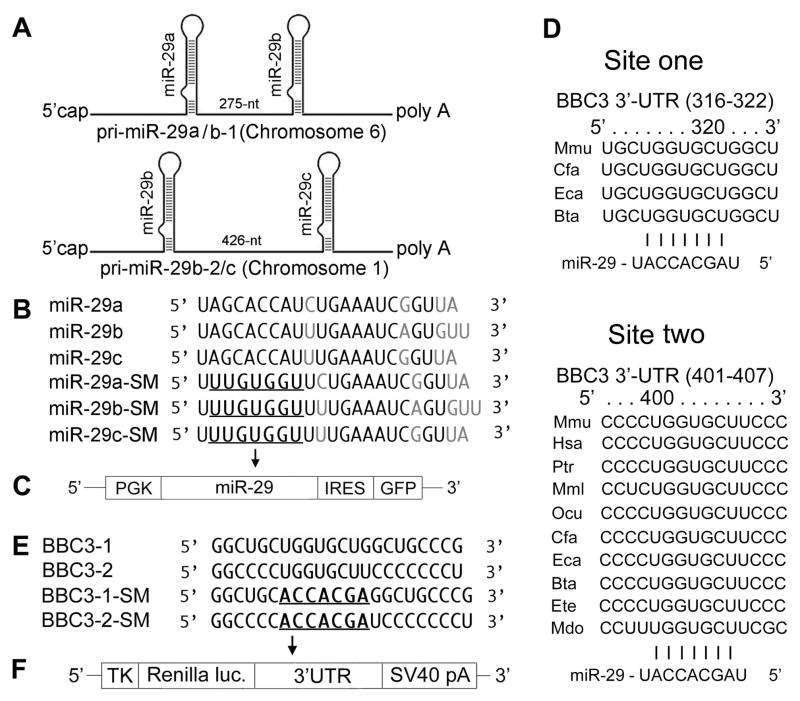
Fig. 1.
A. Schematic representation of the genomic organization of mouse miR-29. The two clusters are on chromosomes 1 and 6. B. Sequences of mature wild type (WT) and seed mutated (SM) miR-29a, miR-29bb and miR-29c. SMs are used as negative controls. C. Vector MWX-PGK-IRES-GFP to express pri-miR-29 and its SM. D. Sequence and alignment of the miR-29-binding sites in the 3′UTR of PUMA. BBC3 has two predicted target sites (from TargetScan): the first (316–322) is less broadly conservative than the second one (401–407). E. Two wild type (BBC3-1 and BBC3-2) and seed mutant (BBC3-1-SM and BBC3-2-SM) in the 3′UTR of PUMA. F. Renilla luciferase reporter vector phRL-TK to express 3′UTRs of PUMA.