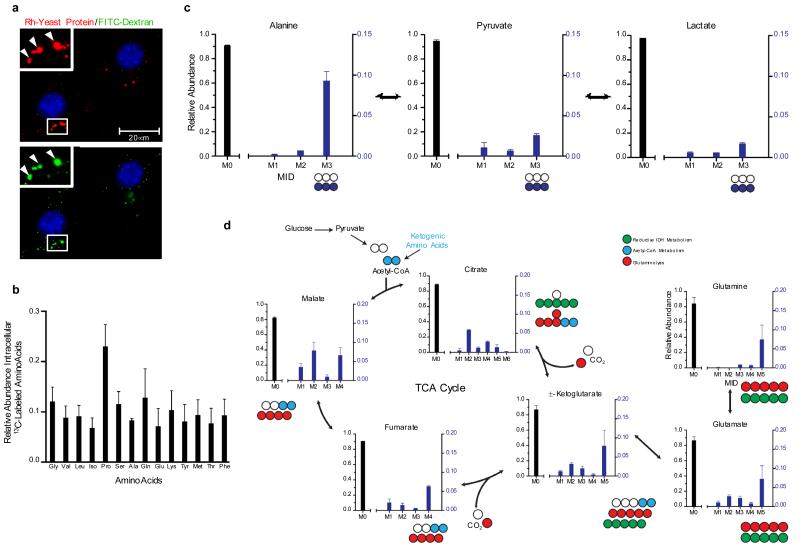
Figure 3. Macropinocytic uptake of extracellular protein drives the accumulation of catabolic intermediates and entry of protein-derived amino acids into central carbon metabolism.
a, Rhodamine-labeled (Rh) yeast protein (red) is internalized into puncta (arrowheads) that co-localize with FITC-dextran (green). Insets represent a higher magnification of the boxed areas. b, Uniformly 13C-labeled intracellular amino acid pools were detected in NIH 3T3 [K-RasV12] cells after culture in low glutamine-containing medium (0.2 mM) supplemented with 2% 13C-labeled yeast protein. c, Protein-derived alanine enters central carbon metabolism upon transamination to pyruvate and pyruvate can be directly converted to lactate. M3 reflects fully labeled alanine, pyruvate and lactate, while M0 abundances reflect metabolites with no 13C label. M1 and M2 represent partially labeled species that are not present in significant amounts. d, Atom transition map depicting a model for the entry of amino acid-derived carbons into the TCA cycle and isotopic labeling of various metabolites. Open circles represent unlabeled carbon, and different colored circles highlight labeling patterns that correspond to specific pathways. For all graphs, error bars indicate mean +/− SD for three independent experiments. IDH, isocitrate dehydrogenase; MID, mass isotopomer distribution