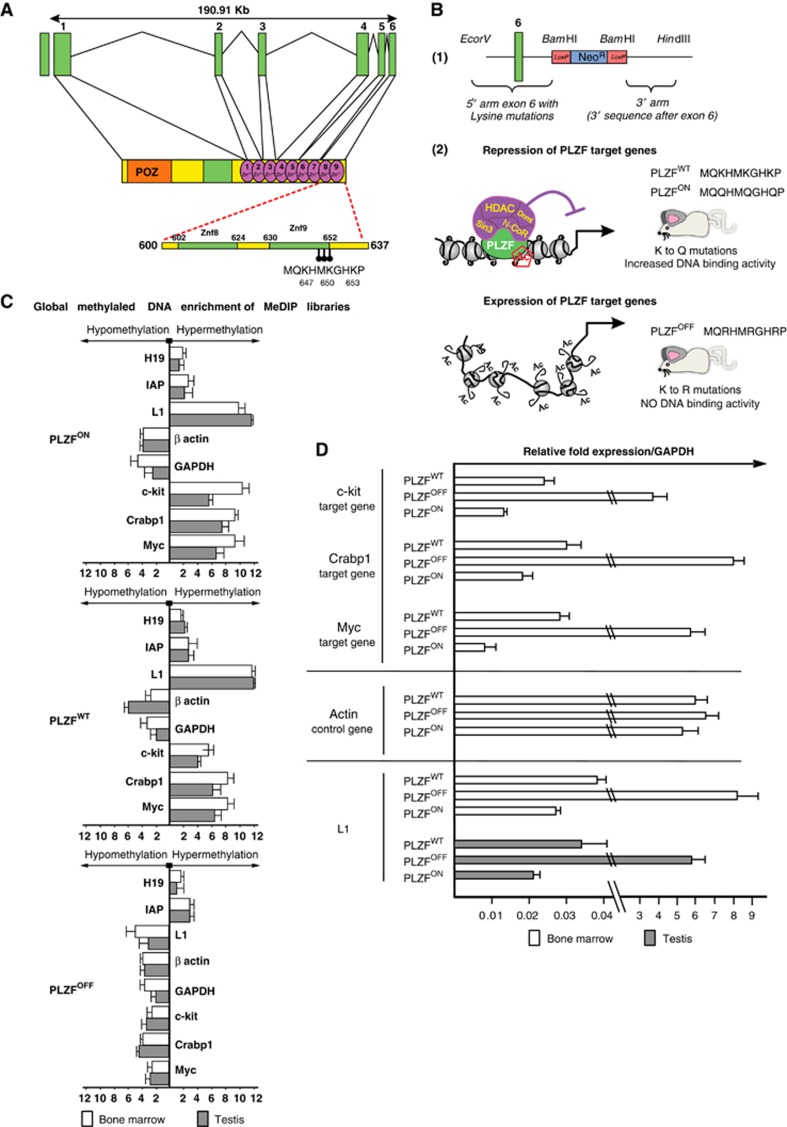
Figure 1.
PLZF-induced DNA methylation throughout the mouse genome. (A) Schematic representation of the structure of the mouse PLZF gene. Green boxes represent exons and connected black straight lines represent intronic regions. The PLZF protein structure is shown under the intronic-exonic organization of the PLZF gene. The orange box represents the POZ domain; the green box represents the proline-rich region; and the green circles represent individual zinc fingers. The acetylation site is situated at the C-terminal end of the most C-terminal zinc finger (9 Zn++), encoded by exon 6, and consists of three lysine residues (K 647, 650 and 653; described in Guidez et al, 2005). These amino acids and their position are shown in the acetylation site sequence. (B) PLZF-targeting vectors and animal models. (1) Schematic representation of the targeting vector pKo scrambler used for the generation of the knock-in embryonic cells. The green box represents exon 6 of the murine PLZF gene, which encodes zinc finger 9 of the PLZF protein. The neomycin cassette is shown in blue and is flanked by two LoxP sites (red boxes), which are used to remove the resistance cassette after recombination. (2) Schematic representation of the impact of the mutant PLZF on chromatin structure in knock-in mice. The PLZFOFF knock-in model expresses a PLZF protein that is unable to bind DNA (due to the mutation of the key lysine residues described above to arginine) and do not recruit co-repressor complex to target sequences; thus, the PLZF-associated genes are constitutively active. The PLZFON knock-in model expresses a PLZF protein that constitutively binds to DNA (due to mutation of the lysine residues in the PLZF acetylation site to glutamine) and recruits like the PLZFWT a co-repressor complex; thus, the PLZF-associated genes are constitutively repressed. (C) Global methylated DNA enrichment of the MeDIP libraries. DNA libraries were prepared from the BM and testis from PLZFOFF, PLZFON and PLZFWT mice. MeDIP (immunoprecipitation of methylated DNA using an antibody raised against 5-methylcytosine (5mC)) was performed to purify genomic methylated DNA regions. The enriched DNA was monitored using quantitative PCR for sequences known to be methylated in mice (the H19 gene and repeated sequences (IAP and L1 elements)) in comparison to input DNAs, as shown in the PLZFWT diagram. Housekeeping genes (GAPDH and beta actin) were used as negative controls. To validate the effect of the PLZF mutants in mice, we performed quantitative PCR of previously described PLZF target genes, including c-kit, Crabp1 and Myc, from the methylation-enriched DNA to determine their levels of DNA methylation (hypomethylation in PLZFOFF samples compared to hypermethylation in PLZFON and PLZFWT samples). (D) Gene expression associated with the presence of mutant or wild-type PLZF proteins. RNAs from mouse tissues were isolated and gene expression was assessed by quantitative PCR using GAPDH expression as a control. Known PLZF targets (c-kit, Myc and Crabp1 genes, A.1) were used to monitor PLZF repressive function while actin served as an internal control (A.2). L1 RNA expression levels were monitored in the bone marrow and testis to correlate the DNA methylation pattern and gene expression of these PLZF-regulated repeats.