Table 1.
Clustering and association analysis results in chromosome 1 simulations
| Method | Cluster | Memb1 | TPR1 | Memb2 | TPR2 | Memb3 | TPR3 | Memb4 | TPR4 | Memb5 | TPR5 | TPR | FP | Time |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A-clustering | ||||||||||||||
| Pearson Correlation | ||||||||||||||
| Average | ||||||||||||||
| d+Aclust (0.15) | 1068.86 | 5.41 | 0.61 | 7.00 | 0.73 | 5.89 | 0.70 | 6.76 | 0.80 | 3.95 | 0.76 | 0.26 | 0.31 | 143.98 |
| Aclust (0.25) | 1241.28 | 3.41 | 0.71 | 7.00 | 0.72 | 5.94 | 0.67 | 7.95 | 0.77 | 4.00 | 0.76 | 0.27 | 0.44 | 273.36 |
| Complete | ||||||||||||||
| d+Aclust (0.15) | 1068.67 | 5.41 | 0.61 | 7.00 | 0.73 | 5.89 | 0.70 | 6.76 | 0.80 | 3.95 | 0.76 | 0.26 | 0.31 | 137.10 |
| Aclust (0.35) | 1656.38 | 3.25 | 0.70 | 7.00 | 0.71 | 5.96 | 0.64 | 8.13 | 0.75 | 4.22 | 0.70 | 0.22 | 0.45 | 138.95 |
| Spearman Correlation | ||||||||||||||
| Average | ||||||||||||||
| d+Aclust (0.20) | 771.60 | 5.91 | 0.65 | 7.00 | 0.76 | 6.00 | 0.73 | 8.06 | 0.82 | 3.98 | 0.80 | 0.32 | 0.44 | 260.21 |
| Aclust (0.30) | 1018.20 | 3.79 | 0.67 | 7.00 | 0.73 | 5.95 | 0.69 | 8.78 | 0.77 | 4.00 | 0.78 | 0.24 | 0.60 | 132.14 |
| Complete | ||||||||||||||
| d+Aclust (0.20) | 771.21 | 5.89 | 0.65 | 7.00 | 0.76 | 5.92 | 0.73 | 8.06 | 0.82 | 3.98 | 0.80 | 0.32 | 0.44 | 299.06 |
| Aclust (0.35) | 1113.85 | 3.23 | 0.66 | 7.00 | 0.73 | 5.82 | 0.70 | 8.32 | 0.78 | 4.15 | 0.77 | 0.24 | 0.60 | 115.35 |
| Bump Hunting | – | 4.49 | 0.59 | 7.00 | 0.69 | 5.99 | 0.62 | 9.49 | 0.76 | 4.00 | 0.42 | 0.11 | 0.23 | 789.48 |
Note: Results of the proposed analysis pipeline based on different parameters of the Aclust algorithm, and the more performant implementation of Bump Hunting. Aclust and d+Aclust stand A-clustering without (Aclust) and with (d+Aclust) 999-dbp-merge initiation step. 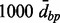 restriction was always applied. The numbers in parentheses are the distance thresholds
restriction was always applied. The numbers in parentheses are the distance thresholds 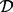 used in each clustering implementation. These thresholds are the optimal ones for each settings, as determined by sensitivity analysis described in the Supplementary Material. Cluster provides the total number of detected clusters by Aclust. Memb m is the mean number of members in the mth cluster, and TPR m is the proportion of simulations in which the mth cluster was found to be significantly associated with the exposure after FDR correction. TPR is the proportion of simulations in which all five clusters were associated with exposure, and FP is the mean number of clusters that were falsely detected as associated with exposure. Time is the elapsed computation time (in seconds).
used in each clustering implementation. These thresholds are the optimal ones for each settings, as determined by sensitivity analysis described in the Supplementary Material. Cluster provides the total number of detected clusters by Aclust. Memb m is the mean number of members in the mth cluster, and TPR m is the proportion of simulations in which the mth cluster was found to be significantly associated with the exposure after FDR correction. TPR is the proportion of simulations in which all five clusters were associated with exposure, and FP is the mean number of clusters that were falsely detected as associated with exposure. Time is the elapsed computation time (in seconds).
