Fig. 1.
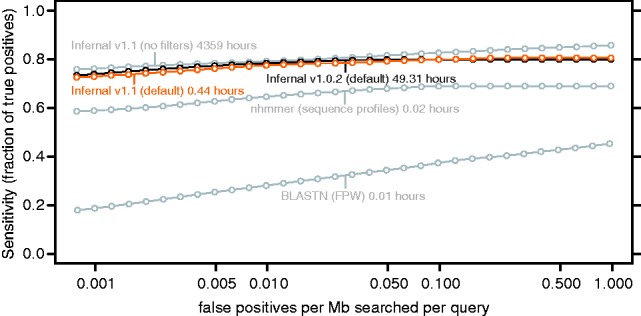
ROC-like curves for the benchmark. Plots are shown for the new Infernal 1.1 with and without filters, for the old Infernal 1.0.2, for profile HMM searches with nhmmer (from the HMMER package included in Infernal 1.1, default parameters) and for family-pairwise-searches with BLASTN (ncbi-blast-2.2.28+, default parameters). The maximum sensitivity (not shown) for default Infernal 1.1 is 0.81 (629 of 820 true positives found), which is achieved at a false-positive rate of 0.19/Mb/query. For non-filtered Infernal, maximum sensitivity is 0.87 at 2.9 false positives per Mb per query. This indicates that at high false-positive rates the filters prevent some true positives from being found, but prevent many more false positives from being found. CPU times are total times for all 106 family searches measured for single execution threads on 3.0 GHz Intel Xeon processors. The Infernal times do not include time required for model calibration.
