Fig. 3.
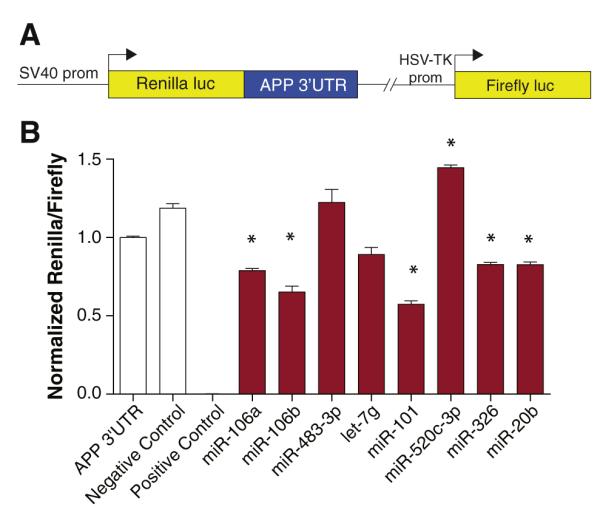
Validation of predicted miRNA targets by reporter assay. (A) An APP 3′-UTR reporter construct was generated as previously described (Long and Lahiri, 2011b). This reporter construct contains the APP 3′-UTR inserted downstream of the Renilla luciferase coding sequence and transcription driven by the SV40 promoter. Expression of firefly luciferase reporter is driven by an independent HSV-TK promoter and serves to normalize for experimental variables such as cell density, transfection efficiency, and cell toxicity. (B) HeLa cells were co-transfected with an APP 3′-UTR reporter construct and miRNAs (50 nM) predicted to target the APP 3′-UTR by various predictor algorithms. Negative control miRNA not predicted to target the APP 3′-UTR was also cotransfected. A positive control was included that consisted of co-transfection with miR-1 and a miR-1 MRE reporter. Renilla and firefly luciferase activity was assayed 48 h post-transfection using the Dual-Glo assay (Promega). Renilla luminescence values are normalized to firefly luminescence and scaled. Statistically significant differences were assessed by ANOVA followed by post-hoc Dunnett’s test. n=6; *p<0.05 relative to negative control miRNA.
