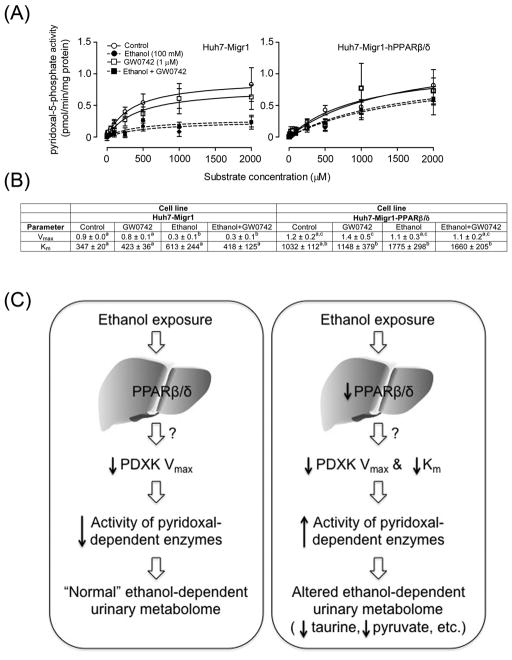
Fig. 8.
Over-expression and ligand activation of PPARβ/δ prevents the ethanol-induced decrease in PDXK Vmax and maintains Km. (A) Michaelis-Menten kinetic parameters were derived from the enzyme assay for PDXK activity using extracts control Huh7 cells (Huh7-Migr1) or Huh7 cells over-expressing PPARβ/δ (Huh-Migr1-hPPARβ/δ) in the presence or absence of ethanol, the highly specific PPARβ/δ ligand GW0742, or both ethanol and GW0742. (B) Vmax and Km values calculated from the enzyme assay for PDXK activity using control Huh7 cells (Huh7-Migr1) or Huh7 cells over-expressing PPARβ/δ (Huh-Migr1-hPPARβ/δ) in the presence or absence of ethanol, the highly specific PPARβ/δ ligand GW0742, or both ethanol and GW0742. (C) Hypothetical model showing how PPARβ/δ alters ethanol metabolism by altering PDXK activity. In normal conditions (left panel), ethanol exposure causes a decrease in hepatic PDXK activity by decreasing Vmax, likely causing decreased activity of a number of pyridoxal- dependent enzymes leading to alterations in the urinary metabolome. In the absence of PPARβ/δ expression and/or the presence of a loss-of-function polymorphism (right panel), ethanol exposure causes an increase in hepatic PDXK activity by markedly decreasing PDXK Km, likely causing increased activity of a number of pyridoxal-dependent enzymes leading to alterations in the urinary metabolome including decreased taurine and pyruvate. These data collectively indicate that PPARβ/δ decreases PDXK activity by maintaining Km in response to ethanol exposure.