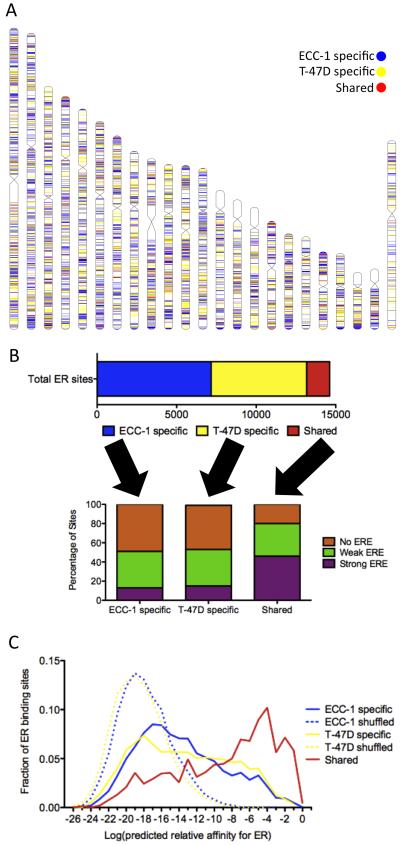
Figure 1. Shared and cell-specific sites have unique sequence properties.
A) The locations of ECC-1 specific (blue), T-47D specific (yellow) and shared (red) ER binding sites are distributed across human chromosomes with an overall lack of shared sites. B) Less than 10% of the total set of identified ER binding sites is consistently bound in ECC-1 and T-47D (top panel). Less than 20% of cell-specific ER binding sites contain EREs, while 46% of shared sites harbor EREs (bottom panel). C) The distribution of the predicted relative affinity for the best match to an ERE is shown for each ECC-1 specific (solid blue), T-47D specific (solid yellow) and shared (solid red) ER binding sites as well as ECC-1 and T-47D ER binding sites in which the order of the nucleotides has been shuffled (dashed lines). All three sets of observed binding sites (solid lines) are enriched over background (dashed lines); however, shared sites represent the highest affinity matches to an ERE. See also Figure S1.