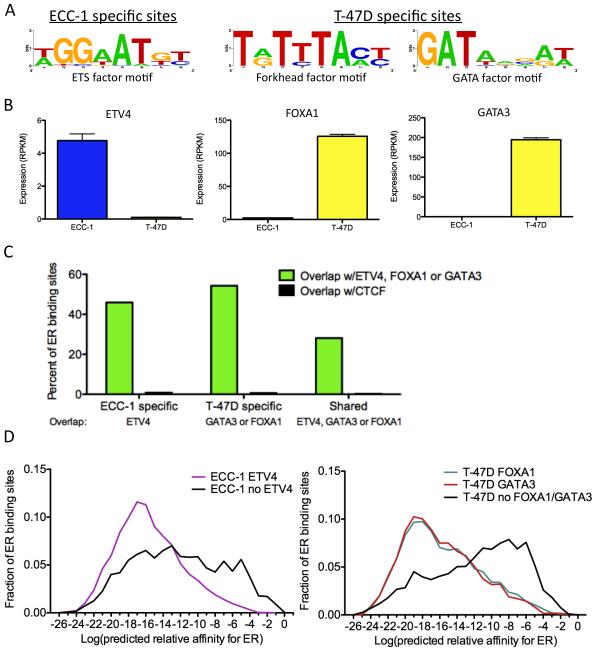
Figure 4. Transcription factors co-occur with ER at cell-specific binding sites.
A) Motifs were identified in ECC-1 (left panel) and T-47D (right panel) ER binding sites after EREs were masked out. B) The expression levels from RNA-seq data of transcription factors that recognize each family of motifs were analyzed and identified ETV4, FOXA1 and GATA3 as potential ER interacting factors. Error bars represent s.e.m. C) Cell-specific ER binding sites overlap ETV4, FOXA1 and GATA3 binding sites measured by ChIP-seq at a higher rate than shared ER binding sites. In each case, the overlap is significantly higher than CTCF, which serves as a negative control. D) ER binding sites that co-occur with these transcription factors in ECC-1 cells (left panel) and T-47D cells (right panel) lack strong matches to an ERE. See also Table S1.