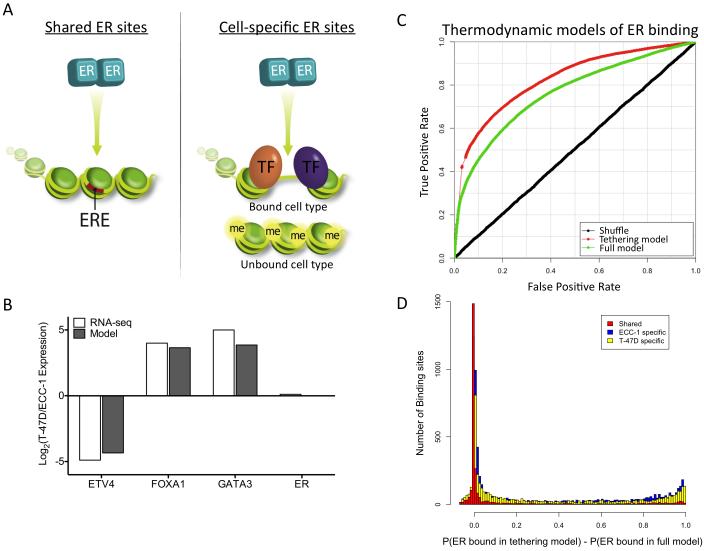
Figure 5. Model of cell type specificity in ER binding site selection.
A) The model depicts the differences between shared and cell-specific binding sites with respect to the presence of EREs (red bars), chromatin accessibility, DNA methylation (“me” yellow circles) and co-occurring transcription factors (“TF” orange and purple ovals). B) Model predictions (gray) of the relative expression of each factor between cell lines are highly correlated with observed transcript levels from RNA-seq experiments (white). C) ROC curves show that the thermodynamic model performs significantly better when a tethering model of ER binding is implemented. D) The distributions of differences between the probability of ER binding in the tethering model and the full model are significantly different between the shared binding sites (red) and either the ECC-1 specific (blue) or T-47D specific (yellow) binding sites. See also Figure S4.