Abstract
The symbiotic bacterium Buchnera aphidicola lacks key genes in the biosynthesis of five essential amino acids (EAAs), and yet its animal hosts (aphids) depend on the symbiosis for the synthesis of these EAAs (isoleucine, leucine, methionine, phenylalanine, and valine). We tested the hypothesis, derived from genome annotation, that the missing Buchnera reactions are mediated by host enzymes, with the exchange of metabolic intermediates between the partners. The specialized host cells bearing Buchnera were separated into a Buchnera fraction and a Buchnera-free host cell fraction (HF). Addition of HF to isolated Buchnera preparations significantly increased the production of leucine and phenylalanine, and recombinant enzymes mediating the final reactions in branched-chain amino acid and phenylalanine synthesis rescued the production of these EAAs by Buchnera preparations without HF. The likely precursors for the missing proximal reactions in isoleucine and methionine synthesis were identified, and they differed from predictions based on genome annotations: synthesis of 2-oxobutanoate, the aphid-derived precursor of isoleucine synthesis, was stimulated by homoserine and not threonine via threonine dehydratase, and production of the homocysteine precursor of methionine was driven by cystathionine, not cysteine, via reversal of the transsulfuration pathway. The evolution of shared metabolic pathways in this symbiosis can be attributed to host compensation for genomic deterioration in the symbiont, involving changes in host gene expression networks to recruit specific enzymes to the host cell.
INTRODUCTION
The capacity of various insect groups to utilize nutritionally unbalanced diets is correlated with possession of vertically transmitted, obligate symbiotic microorganisms with reduced genomes (1–3). Unexpectedly, the genomes of bacterial symbionts of aphids and other sternorrhynchan insects (aphids, mealybugs, whiteflies, and psyllids) are missing genes in the biosynthetic pathways for essential amino acids (EAAs) required by the host (4–6). The pea aphid genome, like other insect genomes, contains genes coding for enzymes with functions equivalent to those of enzymes encoded by genes lost by the obligate bacterial symbionts (7). The proposal that metabolic pathways are shared between the pea aphid Acyrthosiphon pisum and its symbiotic bacterium Buchnera aphidicola was unprecedented (7, 8), as metabolic pathways are traditionally defined as the property of individual organisms.
Buchnera lacks the genes coding for the terminal reaction in the synthesis of both branched-chain amino acids (BCAAs) and phenylalanine and proximal reactions in isoleucine and methionine biosynthesis (4). The terminal reactions for BCAA and phenylalanine synthesis have been predicted to be mediated by host enzymes, branched-chain aminotransferase (BCAT) and an aspartate aminotransferase (GOT2 [glutamate oxaloacetate transaminase 2]), respectively (8). These enzymes are generally present in animals, including insects. Consistent with this expectation, the transcript and protein of the candidate aphid enzymes contributing to these metabolic pathways are enriched in the specialized host cells (bacteriocytes) that solely house the bacterial symbiont, Buchnera (9–11). In contrast to the consensus among genomic, transcriptomic, and proteomic data sets for the enzymes performing the terminal reactions, there are multiple candidate precursors and host enzymes for the proximal reactions contributing to the production of isoleucine and methionine (9, 10). Furthermore, these predictions do not exclude the alternative explanation for the sustained synthesis of EAAs: that the reactions normally catalyzed by the products of genes missing in Buchnera are mediated by other Buchnera enzymes (and not host enzymes) with a greater or different substrate range than indicated by their annotation (4, 12).
Resolution of the question of whether some EAAs are synthesized by shared metabolic pathways or entirely by Buchnera is central to our understanding of the coevolutionary interactions in this relationship because the Buchnera-derived EAAs are required by the aphid to utilize plant phloem sap, a diet grossly deficient in these nutrients (13). This issue is also pertinent for bacterial symbioses in other phloem-feeding insects that have subsequently been invoked to have shared EAA biosynthesis pathways, based on analyses of bacterial gene content (5, 6, 11).
To test the hypothesis of shared EAA biosynthetic pathways in the pea aphid-Buchnera symbiosis, we initiated a study to investigate whether host cell lysates could complement the metabolic deficiencies of Buchnera. Our specific goals were to determine (i) whether the terminal transaminase reactions mediating the synthesis of BCAAs (leucine, isoleucine, and valine) and phenylalanine are localized to the Buchnera or host cells and (ii) the identity of the substrates for candidate host reactions mediating the proximal reactions in isoleucine and methionine synthesis.
MATERIALS AND METHODS
Aphid rearing.
Aphids were reared from a single parthenogenetic female collected from an alfalfa field in Freeville, NY, in June 2009. The aphid line, CWR09/18, was screened by PCR and microscopy for bacterial symbionts and found to contain Buchnera aphidicola and no secondary symbionts. The line was maintained on preflowering Vicia faba cv. Windsor at 20°C with a 16:8 light-dark cycle.
Amino acid release and analysis.
Bacteriocytes were dissected from 7-day-old larval aphids in extraction medium (28 mM glucose, 8.6 mM NaCl, 1 mM MgSO4, 0.1 mM CaCl2, 0.25 M sucrose, 50 mM NaH2PO4, 13 mM K2H2PO4 [pH 7.5]), lysed by pipetting 4 to 6 times, and centrifuged at 1,000 × g for 5 min at 4°C to separate the Buchnera-free supernatant (referred to as the host fraction [HF]) from the pellet containing Buchnera cells. The Buchnera cells in the pellet were quantified by hemocytometer counts and diluted to 4 × 108 Buchnera ml−1 in extraction medium. To initiate the release experiment, 5.5 μl of reaction medium was added to 8 replicate samples of 5.5 μl of bacterial suspension. The standard reaction medium comprised the extraction medium supplemented with glutamate, glutamine, serine, aspartate, and 2-oxobutanoate, each at 2 mM, but 2-oxobutanoate was replaced by 2 mM homoserine, threonine, cystathionine, cysteine, or homocysteine where indicated. For determination of BCAT and phenylalanine transaminase activity in the host fraction, the standard reaction medium was supplemented with 2 mM 4-methyl-2-oxopentanoate or phenylpyruvate and added to HF. At 5-min intervals over 40 min, one tube was centrifuged at 1,000 × g for 70 s, and 11 μl of supernatant was immediately flash frozen in liquid nitrogen and stored at −80°C. The experiments were conducted at 22.5°C and were repeated 3 to 5 times on different occasions using different sets of aphids.
The amino acid content of the supernatant was quantified using the AccQ Tag derivatization kit (Waters) by ultraperformance liquid chromatography (UPLC) with a photodiode array (PDA) detector (Waters Acquity). An equal volume of 40 mM HCl was added to 10 μl of each supernatant. Following incubation on ice for 30 min, the sample was centrifuged at 18,000 × g for 10 min at 4°C, and the supernatant was filtered through a 0.45-μm filter plate (Millipore) by centrifugation at 1,500 × g for 10 min. The filtrate (2.5 μl) was derivatized with AccQ Tag (Waters), by following the manufacturer's protocol, and injected into a Waters Acquity UPLC with a PDA detector and AccQ-Tag Ultra 2.1- by 100-mm column. The gradient was as follows, where A is 10% AccQ-Tag Ultra Eluent A in water and B is AccQ-Tag Ultra Eluent B: 0 to 0.54 min, 99.9% A and 0.1% B; 0.54 to 5.74 min, 90.9% A and 9.1% B; 5.74 to 7.74 min, 78.8% A and 21.2% B; 7.74 to 8.04 min, 40.4% A and 59.6% B; 8.04 to 8.64 min, 10% A and 90% B; 8.05 to 8.64 min, 10% A and 90% B; 8.64 to 8.73 min, 99.9% A and 0.1% B; and 8.73 to 9.50 min, 99.9% A and 0.1% B (linear between each time point). Amino acids were determined by comparison to standards: 1, 5, 10, 50, and 100 pmol of amino acids μl−1 (Waters amino acid hydrolysate standard 088122, supplemented with asparagine, tryptophan, and glutamine).
Immunoblots.
Custom-made polyclonal antibodies against glutamate oxaloacetate transaminase 2 (GOT2; NCBI gene identity [ID], 100144899; predicted size, 48 kDa), branched-chain aminotransferase (BCAT; NCBI gene ID, 100167587; predicted size, 49 kDa), and threonine dehydratase (TD; NCBI gene ID, 100165866; predicted size, 40 kDa) were produced by GenScript (Piscataway, NJ). Rabbits were immunized four times using the purified peptides CNPTGVDPKPEQWKE (GOT2), CVDERPHLYESQNYK (BCAT), and CMEHGSPITVDGKST (TD) conjugated with keyhole limpet hemocyanin (KLH). The polyclonal antiserum obtained from the last bleed was affinity purified and tested by enzyme-linked immunosorbent assay (ELISA), including confirmation that each preimmune serum did not react to aphid proteins. The specificity of the antibodies was confirmed using mass spectrometry (MS). Briefly, the reacting band in the Western blot was manually excised from the corresponding acrylamide gel, digested, and submitted to a high-performance liquid chromatography (HPLC) system (Dionex Ultimate 300 configured for nanobore), in-line connected to a hybrid triple quadrupole linear ion trap mass spectrometer, 4000 Q Trap (ABI/MDS Sciex) equipped with an ion spray head for an ion source. The resulting MS data were submitted for database searching using the MASCOT search engine, version 2.3, against a database containing the pea aphid genome with 34,834 protein-coding gene models (www.AphidBase.com).
For protein analysis, whole bodies and dissected bacteriocytes of 7-day-old larval aphids were homogenized in ice-cold buffer containing 35 mM Tris, 25 mM KCl, and 10 mM MgCl2, pH 7.4. Following centrifugation at 10,600 × g for 5 min, the protein content of the supernatant was quantified using the RC DC protein assay kit (Bio-Rad, Hercules, CA) according to the manufacturer's instructions, with bovine serum albumin as a standard. SDS-PAGE (14) was performed with 2.5 μg of protein per sample on a 12% polyacrylamide gel containing 0.1% SDS. For Western blotting, the proteins were transferred to a nitrocellulose membrane and blocked in phosphate-buffered saline (PBS) containing 0.5% Tween (PBS-T) and 5% milk powder. The membrane was incubated successively in PBS-Tween containing a polyclonal antibody diluted to either 1/200 (BCAT and GOT2) or 1/1,000 (TD), with an anti-rabbit IgG conjugated to peroxidase (Sigma) diluted 1/20,000, and then ECL substrate (Bio-Rad), followed by visualization with Molecular Imager ChemiDoc XRS (Bio-Rad). The intensity of the bands was analyzed by Quantity One software (Bio-Rad).
Generation of recombinant enzymes.
Escherichia coli was used as a source of enzymes that mediate the terminal reactions in BCAA synthesis (IlvE), phenylalanine synthesis (TyrB), and the synthesis of a homocysteine precursor of methionine (MetC). The genes were amplified from E. coli JM109 genomic DNA with the following primers: ilvE_forward, 5′-TTTAGGATCCACCACGAAGAAAGCTG-3′; ilvE_ reverse, 5′-TTTAGGTACCGTGTCTGTCTCGTAAA-3′; tyrB_forward, 5′-TTTAGGATCCCAAAAAAGTTGACGCCT-3′; tyrB_reverse, 5′-TTTAGGTACCTTACATCACCGCAGCA-3′; metC_forward, 5′-TTTAGGATCCGCGGACAAAAAGCTTG-3′; and metC_reverse, 5′-TTTAGGTACCTTATACAATTCGCGCAA-3′. Each forward primer includes a BamHI site, and each reverse primer a KpnI site, for ligation into the expression vector pProExHT to yield p6His-IlvE, p6His-TyrB, and p6His-MetC. Successful plasmid construction was confirmed by restriction digests and Sanger sequencing.
Recombinant enzyme was expressed and purified from E. coli BL21 cells, essentially as described previously (15). Briefly, 1 ml of overnight culture was added to 350 ml of LB and grown with shaking at 120 rpm and 37°C for 4.5 h. After 4.5 h, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a concentration of 0.5 mM and the culture was shifted to 25°C for an additional 18 to 20 h of incubation. Cells were pelleted, flash frozen, and stored at −80°C until enzyme purification.
For purification, 5 ml of equilibration buffer (20 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 7.4]) and 50 μl of Halt protease inhibitor cocktail (Thermo Scientific) were added to the frozen cell pellet, and cells were lysed by sonication. Cellular debris was removed by centrifugation at 4,500 × g for 10 min at 4°C. The supernatant was added to 1 ml of equilibrated HisPur nickel-nitrilotriacetic acid (Ni-NTA) resin (Thermo Scientific) and incubated at 4°C for 1.5 h with gentle rocking to allow binding of the His-tagged protein. The resin was then washed 5 times with 5 ml of wash buffer (20 mM NaH2PO4, 300 mM NaCl, 25 mM imidazole [pH 7.4]) for 10 min at 4°C. Resin-bound protein was eluted in three steps with elution buffer (20 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole [pH 7.4]), and the third elution was dialyzed overnight at 4°C in a 7,000-molecular-weight-cutoff (MWCO) membrane against 1 liter of glucose-free extraction buffer (see above). The presence of pure protein was confirmed by SDS-PAGE, and enzymatic activity was demonstrated by incubation with substrate and detection of the predicted product by UPLC (IlvE, isoleucine, leucine, and valine from the cognate oxo acid; TyrB, phenylalanine from phenylpyruvate; and MetC, homocysteine from cystathionine).
To quantify the effect of recombinant enzyme on amino acid production by Buchnera, the reaction medium (described above) was supplemented with 0.2 mM pyridoxal-5-phosphate and 20 μg of recombinant enzyme protein ml−1. The reaction medium for MetC experiments also contained 2 mM cystathionine.
Statistical analyses.
The release rate of amino acids from Buchnera preparations was quantified from the regression of amino acid content in the medium over time. The variation in release rates with treatment was analyzed by analysis of variance (ANOVA), t test, or paired t test, following logarithmic transformation to obtain normally distributions (Shapiro-Wilk test) with homogenous variance (Levene's test), with the least significant difference (LSD) post hoc to determine significant pairwise differences within the ANOVA. Data that failed Levene's test were analyzed with a Kruskal-Wallis test, with pairwise differences determined with Mann-Whitney U tests (P values determined by a Bonferroni correction).
RESULTS
Terminal transamination reactions in synthesis of leucine and phenylalanine.
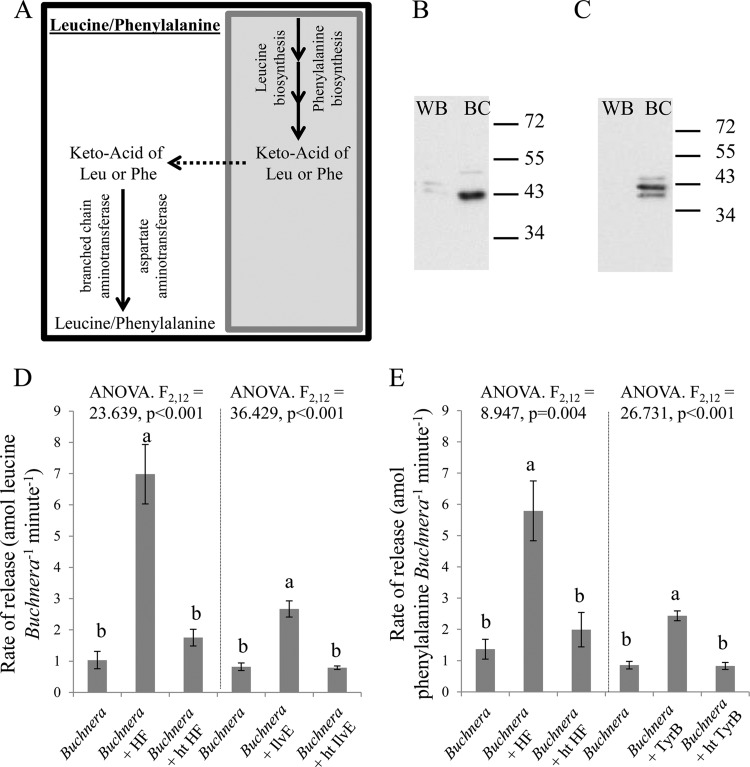
Buchnera lacks recognizable genes that mediate the terminal reactions in the synthesis of phenylalanine and BCAAs in related bacteria (4). The aphid enzymes GOT2 (which is annotated with phenylalanine aminotransferase activity) and BCAT have been hypothesized to mediate the missing reactions in phenylalanine and BCAA biosynthesis, respectively (Fig. 1A) (8–11).
Fig 1.
Leucine and phenylalanine release by isolated Buchnera preparations. (A) Metabolic map of the terminal reactions for leucine and phenylalanine biosynthesis (shaded box represents Buchnera). (B) Western blot of 5 μg of protein from whole aphid (WB) and bacteriocytes (BC) using polyclonal antibody against BCAT. (C) Western blot of 5 μg of protein from WB and BC using polyclonal antibodies against GOT2. (For panels B and C, molecular masses are shown to the right [34 to 72 kDa].) (D) Leucine release from Buchnera with and without HF and release from Buchnera with and without recombinant IlvE. (E) Phenylalanine release from Buchnera with and without HF and release from Buchnera with and without recombinanat TyrB. ht, heat treated. Values are means ± SEs. P < 0.01 by LSD post hoc test (different letters refer to significantly different mean values).
The BCAT and GOT2 proteins were detected in the bacteriocyte when tested by immunoblotting with polyclonal antibodies (Fig. 1B and C). If these soluble enzymes contribute to BCAA and phenylalanine synthesis, then the production of these amino acids by Buchnera preparations is predicted to be stimulated by HF. (Buchnera preparations without HF are expected to produce these EAAs at low rates because they cannot be separated from all contaminating aphid protein, including BCAT and GOT2 [10].) Consistent with these expectations, leucine and phenylalanine are released at low rates from preparations incubated without HF, and the release of these amino acids is significantly elevated by a heat-sensitive component of HF (Fig. 1D and E).
As a complementary approach to investigate the source of the terminal reactions in BCAA and phenylalanine synthesis, we investigated whether the production of these EAAs by HF-free Buchnera can be restored by the addition of recombinant transaminases (IlvE and TyrB). Consistent with the predicted role of host enzyme-mediated transamination, these treatments resulted in significant increases in the release rate of leucine and phenylalanine (Fig. 1D and E). Furthermore, HF was also tested for BCAT and phenylalanine aminotransferase activities. Specifically, HF showed a significant increase in leucine and phenylalanine production (9-fold and 135-fold, respectively) when the medium was supplemented with either 4-methyl-2-oxopentanoate or phenylpyruvate, and this activity was lost when the HF was heat treated (Fig. 2).
Fig 2.
Enzymatic activities in HF. (A) Production of leucine by HF with substrate 4-methyl-2-oxopentanoate. (B) Production of phenylalanine by HF with substrate phenylpyruvate. The three treatments in each experiment are HF with substrate (HF), HF minus substrate, and heat-treated HF with substrate (htHF).
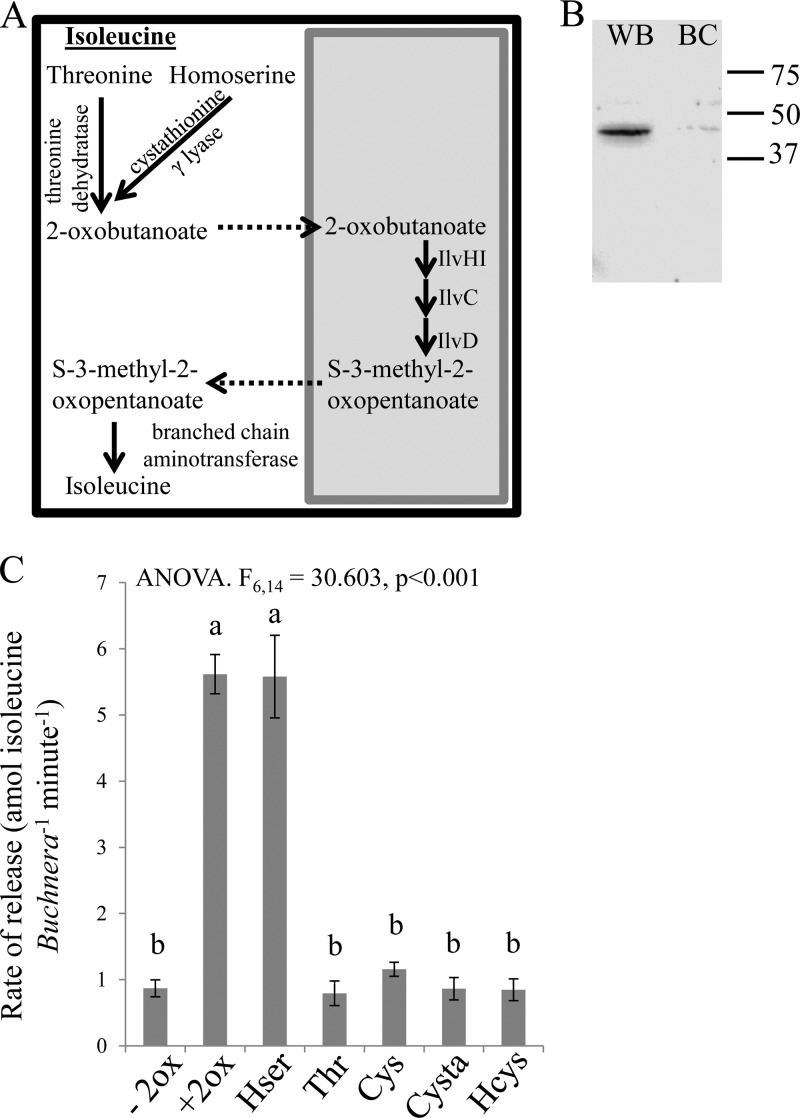
Metabolic source of 2-oxobutanoate, the precursor of isoleucine.
Production of isoleucine is an essential function of the aphid Buchnera symbiosis, yet the source of the key intermediate, 2-oxobutanoate, remains to be defined. The genome reconstruction concluded that the aphid threonine dehydratase transforms threonine to 2-oxobutanoate, with the expectation of threonine dehydratase protein in aphid bacteriocytes (Fig. 3A) (8). Contrary to this prediction, we found that threonine dehydratase was undetectable in the bacteriocytes, although it was readily detected in aphid whole bodies (Fig. 3B). An alternative source of 2-oxobutanoate could be homoserine by cystathionine-γ-lyase, a possibility supported by the enrichment of cystathionine-γ-lyase in the bacteriocyte proteome (10).
Fig 3.
Isoleucine release by preparations of Buchnera with HF. (A) Metabolic map illustrating the potential sources for 2-oxobutanoate based upon candidate enzymes: threonine and homoserine (shaded box represents Buchnera). (B) Western blot of 5 μg of protein from whole aphid (WB) and bacteriocytes (BC) using polyclonal antibody against threonine dehydratase. (C) Isoleucine release from Buchnera plus HF incubated with different precursors: −2ox, minus 2-oxobutanoate (no substrate); +2ox, 2-oxobutanoate; Hser, homoserine; Thr, threonine; Cys, cysteine; Cysta, cystathionine; Hcys, homocysteine. Values are means ± SEs. P < 0.001 by LSD post hoc test.
To investigate the metabolic source of 2-oxobutanoate, Buchnera preparations with HF were supplemented with 2-oxobutanoate or metabolically related compounds (Fig. 3C). Only 2-oxobutanoate and homoserine supported a significantly elevated release rate of isoleucine, compared to medium with no source of 2-oxobutanoate. These findings suggest that the precursor of 2-oxobutanoate is homoserine and not threonine, which did not stimulate isoleucine release.
Metabolic source of homocysteine, the precursor of methionine.
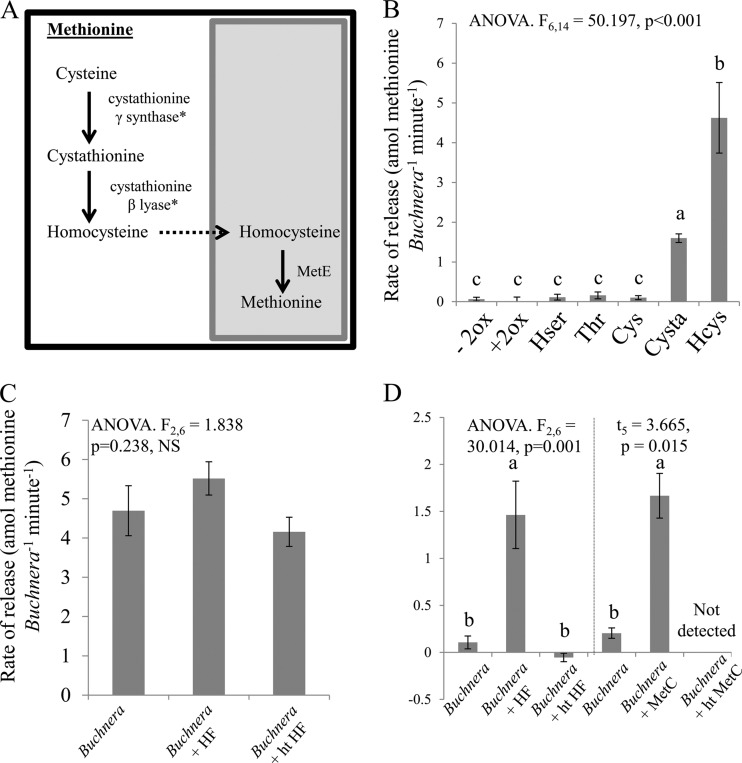
The immediate precursor for methionine synthesis is homocysteine, to which a methyl group is added to form methionine. Buchnera possesses the gene for the terminal enzyme, MetE, but lacks genes for the other steps in methionine biosynthesis. As a result, Buchnera is predicted to require an exogenous supply of homocysteine to support methionine production. It has been hypothesized that the host provides Buchnera with homocysteine by a reversal of the transsulfuration pathway, utilizing cystathionine-γ-lyase (as a cystathionine-γ-synthase) and cystathionine-β-synthase (as a cystathionine-β-lyase) to form cystathionine from cysteine and homocysteine from cystathionine, respectively (Fig. 4A) (8, 9). The reversal of the transsulfuration pathway in animals has never been experimentally shown in any system to date.
Fig 4.
Methionine release by isolated Buchnera preparations. (A) Metabolic map of the reversal of the transsulfuration pathway to produce homocysteine. The shaded box represents Buchnera. The items marked with asterisks were annotated as cystathionine-γ-lyase and cystathionine-β-synthase. (B) Methionine release from Buchnera plus HF incubated with different precursors: −2ox, minus 2-oxobutanoate (no substrate); +2ox, 2-oxobutanoate; Hser, homoserine; Thr, threonine; Cys, cysteine; Cysta, cystathionine; Hcys, homocysteine. (C) Methionine release from Buchnera with and without HF in homocysteine medium. (D) Methionine release from Buchnera with and without HF in cystathionine medium and methionine release from Buchnera with and without recombinant MetC. ht, heat treated. Values are means ± SEs. P < 0.01 by LSD post hoc test.
To investigate the source of homocysteine, Buchnera preparations with HF were incubated with homocysteine or one of several possible precursors for homocysteine. As expected, homocysteine promoted methionine production by Buchnera (73-fold increase over the medium-only control), yet only cystathionine, and not cysteine, promoted methionine production (25-fold increase versus no increase [Fig. 4B]). These findings confirm that aphid homocysteine is the precursor for Buchnera-mediated methionine synthesis but are inconsistent with the prediction that the transsulfuration pathway functions in reverse (8), with methionine ultimately being produced from cysteine. Instead, they suggest that cystathionine is the metabolic precursor for methionine.
To investigate the contribution of host enzymes to methionine synthesis further, Buchnera preparations were supplemented with homocysteine or cystathionine and incubated with or without HF, and the rate of production of methionine was measured. No difference in methionine production was obtained between Buchnera preparations in homocysteine-supplemented medium incubated with and without HF (Fig. 4C), as predicted with the terminal enzyme being a Buchnera enzyme. In cystathionine-supplemented medium, however, the methionine production by Buchnera was significantly elevated by HF addition (14-fold increase [Fig. 4D]). To test whether the enzymatic activity generating homocysteine is external to the bacterium, Buchnera preparations were incubated with recombinant MetC (converts cystathionine to homocysteine). The Buchnera treated with MetC and cystathionine showed a significant increase in methionine production (8-fold increase) compared to that in enzyme-free medium (Fig. 4D). Taken together, these findings indicate that homocysteine can be produced from exogenous cystathionine and can be converted to methionine by Buchnera MetE.
DISCUSSION
Linked to its small gene content, Buchnera is nutritionally fastidious. Buchnera growth is inferred to require 33 host-derived metabolites (11, 16), and there is no reasonable prospect of culturing this bacterium. Despite this, viable and metabolically active Buchnera cells persist in defined medium for some hours (17–19), facilitating direct investigation of their metabolic capabilities. In this study, we probed the metabolic functions of isolated Buchnera preparations and obtained direct evidence that metabolic pathways are shared between host and symbiont. Specifically, we demonstrated (i) net production of leucine and phenylalanine by Buchnera supplemented with either recombinant enzyme mediating the terminal biosynthetic reaction (IlvE and TyrB, respectively) or HF containing functionally equivalent aphid enzymes and (ii) that Buchnera preparations require the exogenous supply of either the substrate or product of host reactions for production of isoleucine and methionine, indicating the host's compensation for missing Buchnera IlvA and MetC.
In addition to validating the host-mediated terminal reactions in leucine and phenylalanine synthesis, this study provides metabolism-based identification of the substrates utilized by the host-mediated proximal reactions. We conclude that the host-derived 2-oxobutanoate precursor of isoleucine synthesis is synthesized from homoserine and not threonine. This runs contrary to the prediction from the aphid genome annotation (8) but is fully congruent with metabolic evidence that pea aphids do not metabolize dietary [14C]threonine to isoleucine (20), and cystathionine-γ-lyase, an enzyme that is enriched in the pea aphid bacteriocytes, has previously been suggested to mediate the synthesis of 2-oxobutanoate from homoserine (10).
Also contrary to predictions based on pathway metabolic pathway reconstruction (8), our results suggest that the homocysteine substrate for Buchnera-mediated methionine synthesis may be derived from cystathionine and not cysteine. The predicted host enzyme mediating homocysteine synthesis from cystathionine, cystathionine-β-synthase (NCBI gene ID, 100166111), is undetectable in aphid bacteriocytes (10), and the enzyme mediating this reaction remains to be identified; detailed analysis of the bacteriocyte proteome has identified no strong candidates (C. W. Russell, unpublished data).
An important consideration in interpreting experiments on isolated symbiotic microorganisms is their relevance to function in the symbiosis. When certain symbionts are separated from their hosts, key symbiotic traits, notably the selective release of nutrients advantageous to the host, are abolished (21). Several lines of evidence suggest that the isolated Buchnera preparations retain symbiosis-relevant metabolic properties. In particular, the sustained release of the penultimate metabolites in the BCAA and phenylalanine biosynthesis pathways can be inferred with confidence because these EAAs are produced by Buchnera preparations incubated with recombinant enzymes mediating the terminal reactions, but an exogenous supply of these precursors is required for the EAA production by Buchnera-free HF. The isolated Buchnera cells apparently do not release the substrate of the proximal host reactions for isoleucine and methionine synthesis (homoserine and cystathionine, respectively), suggesting either that these substrates are derived from the host or that their release from Buchnera cells is lost on isolation. Of these two compounds, Buchnera has the genetic capacity to synthesize only homoserine (via the threonine biosynthetic pathway) and does not have the genes required to make cystathionine. Another consideration is the transport of these compounds between the Buchnera and the pea aphid. The pea aphid has an expanded set of amino acid transporters that could fulfill the role of transport of intermediates (22), yet Buchnera has relatively few transporters of uncertain specificity (4). Future studies will need to be performed to determine the transporters responsible for the flux of the many metabolites between the Buchnera cells and surrounding host cell.
Metabolic cooperation occurs widely among microorganisms, generally in response to selection imposed by resource-poor habitats (23–25). The coevolution of shared metabolic pathways in the aphid-Buchnera symbiosis most probably has a different evolutionary basis. Because the vertically transmitted Buchnera cells have a small effective population size, they are subject to gene loss through genomic deterioration (26, 27), selecting for evolutionary changes in host gene expression networks to recruit enzymes that compensate for the missing bacterial enzymatic reactions in the host cell.
The incidence of shared metabolic pathways (i.e., metabolic pathways in which host and symbiont contribute different enzymatic reactions) in animal-microbial symbioses is largely unstudied but may be widespread, albeit not universal, in associations involving microorganisms with reduced genomes. For example, independently evolved bacterial symbionts in phloem-feeding whiteflies and mealybugs of the same suborder as the aphids, the Sternorrhyncha, have incomplete genetic capacity for EAA biosynthesis (6, 28, 29), but the biosynthetic pathways are apparently complete in the bacterial symbionts, e.g., Sulcia, of insects (e.g., cicadas, spittlebugs, and leafhoppers) in the related suborder, the Auchenorryncha (30–32). Turning to different symbioses, the stimulation of EAA production from Buchnera preparations by the host cell fraction (HF) is reminiscent of enhanced photosynthate release by symbiotic algae isolated from corals or other marine invertebrates and incubated with homogenates of their hosts (33). The similarity may, however, be superficial because the effect of host homogenate on algal cells is generally interpreted as a chemical signal that induces the synthesis and export of specific photosynthesis-derived nutrients (34, 35).
In conclusion, the shared metabolic pathways in the pea aphid-Buchnera symbiosis is the product of metabolic coevolution, involving the loss of Buchnera genes mediating reactions also present in animals, as well as the compensatory enrichment of expression of the animal enzymes in host cells housing the symbionts. Associated coevolutionary changes in the regulation of flux through the metabolic pathways (e.g., by feedback inhibition) or in host and symbiont transporters mediating the transfer of metabolic intermediates between the partners are also anticipated (22). Importantly, the host enzymes implicated in symbiotic EAA biosynthesis are localized to the host cell (10) and not derived by lateral gene transfer from the ancestral Buchnera or other bacteria (36). In this respect, the evolutionary trajectory of these symbionts displaying metabolic coevolution differs markedly from that of the bacterium-derived organelles whose metabolism is sustained by the products of organelle-derived genes transferred to the host nucleus (2).
ACKNOWLEDGMENTS
We thank H. Sondermann (Cornell University), who provided the expression vector pProExHT, and colleagues in the Cornell University Life Science Core Proteomics and Mass Spectrometry facility for conducting the proteomics analysis.
Financial support was provided by the National Science Foundation, USA (IOS-0919765, to A.E.D.) and a graduate student fellowship from Sarkaria Institute of Insect Physiology and Toxicology (to C.W.R.).
Footnotes
Published ahead of print 26 July 2013
REFERENCES
- 1.Buchner P. 1965. Endosymbioses of animals with plant microorganisms. John Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 2.McCutcheon JP. 2010. The bacterial essence of tiny symbiont genomes. Curr. Opin. Microbiol. 13:73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shigenobu S, Wilson ACC. 2011. Genomic revelations of a mutualism: the pea aphid and its obligate bacterial symbiont. Cell. Mol. Life Sci. 68:1297–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86 [DOI] [PubMed] [Google Scholar]
- 5.McCutcheon JP, von Dohlen CD. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 21:1366–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sloan DB, Moran NA. 2012. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol. Lett. 8:986–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Aphid Genomics Consortium 2010. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313. 10.1371/journal.pbio.1000313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson AC, Ashton PD, Calevro F, Charles H, Colella S, Febvay G, Jander G, Kushlan PF, Macdonald SJ, Schwartz JF, Thomas GH, Douglas AE. 2010. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol. Biol. 19(Suppl 2::249–258 [DOI] [PubMed] [Google Scholar]
- 9.Hansen AK, Moran NA. 2011. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc. Natl. Acad. Sci. U. S. A. 108:2849–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poliakov A, Russell CW, Ponnala L, Hoops HJ, Sun Q, Douglas AE, van Wijk KJ. 2011. Large-scale label-free quantitative proteomics of the pea aphid-Buchnera symbiosis. Mol. Cell. Proteomics 10:M110.007039. 10.1074/mcp.M110.007039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macdonald SJ, Lin GG, Russell CW, Thomas GH, Douglas AE. 2012. The central role of the host cell in symbiotic nitrogen metabolism. Proc. Biol. Sci. 279:2965–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelkar YD, Ochman H. 2013. Genome reduction promotes increase in protein functional complexity in bacteria. Genetics 193:303–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akman Gündüz E, Douglas AE. 2009. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc. Biol. Sci. 276:987–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 15.Newell PD, Boyd CD, Sondermann H, O'Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 9:e1000587. 10.1371/journal.pbio.1000587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacDonald SJ, Thomas GH, Douglas AE. 2011. Genetic and metabolic determinants of nutritional phenotype in an insect-bacterial symbiosis. Mol. Ecol. 20:2073–2084 [DOI] [PubMed] [Google Scholar]
- 17.Douglas AE, Bouvaine S, Russell RR. 2011. How the insect immune system interacts with an obligate symbiotic bacterium. Proc. Biol. Sci. 278:333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki T, Ishikawa H. 1995. Production of essential amino acids from glutamate by mycetocyte symbionts of the pea aphid, Acyrthosiphon pisum. J. Insect Physiol. 41:41–46 [Google Scholar]
- 19.Whitehead LF, Douglas AE. 1993. A metabolic study of Buchnera, the intracellular bacterial symbionts of the pea aphid, Acyrthosiphon pisum. J. Gen. Microbiol. 139:821–826 [Google Scholar]
- 20.Febvay G, Liadouze I, Guillaud J, Bonnot G. 1995. Analysis of energetic amino acid metabolism in Acyrthosiphon pisum—a multi-dimensional approach to amino acid metabolism in aphids. Arch. Insect Biochem. Physiol. 29:45–69 [Google Scholar]
- 21.Douglas AE. 2010. The symbiotic habit. Princeton University Press, Princeton, NJ [Google Scholar]
- 22.Price DR, Duncan RP, Shigenobu S, Wilson AC. 2011. Genome expansion and differential expression of amino acid transporters at the aphid/Buchnera symbiotic interface. Mol. Biol. Evol. 28:3113–3126 [DOI] [PubMed] [Google Scholar]
- 23.Wintermute EH, Silver PA. 2010. Dynamics in the mixed microbial concourse. Genes Dev. 24:2603–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klitgord N, Segre D. 2011. Ecosystems biology of microbial metabolism. Curr. Opin. Biotechnol. 22:541–546 [DOI] [PubMed] [Google Scholar]
- 25.Freilich S, Zarecki R, Eilam O, Segal ES, Henry CS, Kupiec M, Gophna U, Sharan R, Ruppin E. 2011. Competitive and cooperative metabolic interactions in bacterial communities. Nat. Commun. 2:589. [DOI] [PubMed] [Google Scholar]
- 26.Moran NA. 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. U. S. A. 93:2873–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moran NA, Mira A. 2001. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2:R0054. 10.1186/gb-2001-2-12-research0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabree ZL, Huang CY, Okusu A, Moran NA, Normark BB. 2013. The nutrient supplying capabilities of Uzinura, an endosymbiont of armoured scale insects. Environ. Microbiol. 15:1988–1999 [DOI] [PubMed] [Google Scholar]
- 29.Jiang Z-H, Xia F, Johnson KW, Brown CD, Bartom E, Tuteja JH, Stevens R, Grossman RL, Brumin M, White KP, Ghanim M. 2013. Comparison of the genome sequences of “Candidatus Portiera aleyrodidarum” primary endosymbionts from the whitefly Bemisia tabaci B and Q biotypes. Appl. Environ. Microbiol. 79:1757–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCutcheon JP, McDonald BR, Moran NA. 2009. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl. Acad. Sci. U. S. A. 106:15394–15399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCutcheon JP, Moran NA. 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. U. S. A. 104:19392–19397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCutcheon JP, Moran NA. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol. Evol. 2:708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trench RK. 1971. Physiology and biochemistry of zooxanthellae symbiotic with marine coelenterates. III. Effect of homogenates of host tissues on excretion of photosynthetic products in vitro by zooxanthellae from two marine coelenterates. Proc. Biol. Sci. 177:251–269 [Google Scholar]
- 34.Grant AJ, Trautman DA, Menz I, Hinde R. 2006. Separation of two cell signalling molecules from a symbiotic sponge that modify algal carbon metabolism. Biochem. Biophys. Res. Commun. 348:92–98 [DOI] [PubMed] [Google Scholar]
- 35.Wang JT, Douglas AE. 1997. Nutrients, signals, and photosynthate release by symbiotic algae. Plant Physiol. 114:631–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikoh N, McCutcheon JP, Kudo T, Miyagishima SY, Moran NA, Nakabachi A. 2010. Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet. 6:e1000827. 10.1371/journal.pgen.1000827 [DOI] [PMC free article] [PubMed] [Google Scholar]