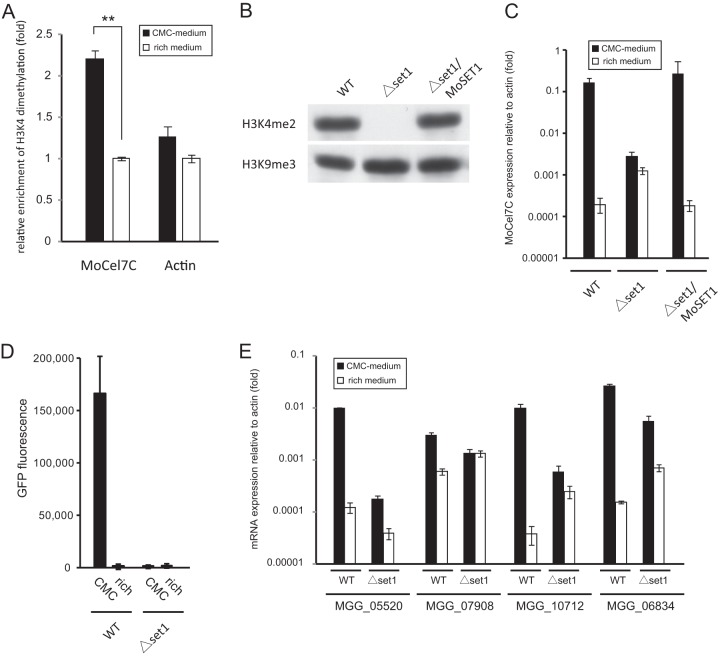
Fig 4.
(A) Chromatin immunoprecipitation (ChIP) analysis of H3K4me2 levels at the MoCel7C and actin genes. The analysis was performed 36 h after transfer to medium containing 1% carboxymethylcellulose (CMC) (black bars) or conventional rich medium (white bars). Immunoprecipitated and input DNA were quantified by qPCR. ChIP signal was normalized to input DNA. Relative enrichment of H3K4me2 under inducing conditions was calculated by dividing normalized data by those under noninducing conditions. Error bars represent standard deviations. **, significant difference (two-tailed t test, P < 0.01). This experiment was performed with two biological replicates with duplicate technical replicates. (B) Western blot analysis of H3K4me2 in the MoSET1 KO strain. Total protein extracted from the wild type (WT), the MoSET1 KO mutant (Δset1), and the complementary strain (Δset1/MoSET1) was separated on a 15% polyacrylamide gel and probed with rabbit polyclonal antibodies against H3K4me2 (active motif). (C) qRT-PCR analysis of MoCel7C expression in response to CMC. The WT, Δset1, and Δset1/MoSET1 strains were grown on medium containing 1% CMC or on conventional rich medium for 36 h. Values are relative expression levels of the MoCel7C gene relative to that of the actin gene (set at 1). This experiment was performed with two biological replicates and duplicate technical replicates. Error bars represent standard deviations. (D) GFP reporter assay of the MoCel7C promoter in response to CMC. Two pEGEX2-GFP transformants each of the WT and Δset1 strains were grown on medium containing 1% CMC for 48 h and subjected to GFP measurement. Error bars represent standard deviations. This experiment was performed with three technical replicates. (E) qRT-PCR analysis of the CMC-responsive cellulase genes in the MoSET1 KO (Δset1) mutant. Experiments were performed as described for panel C.