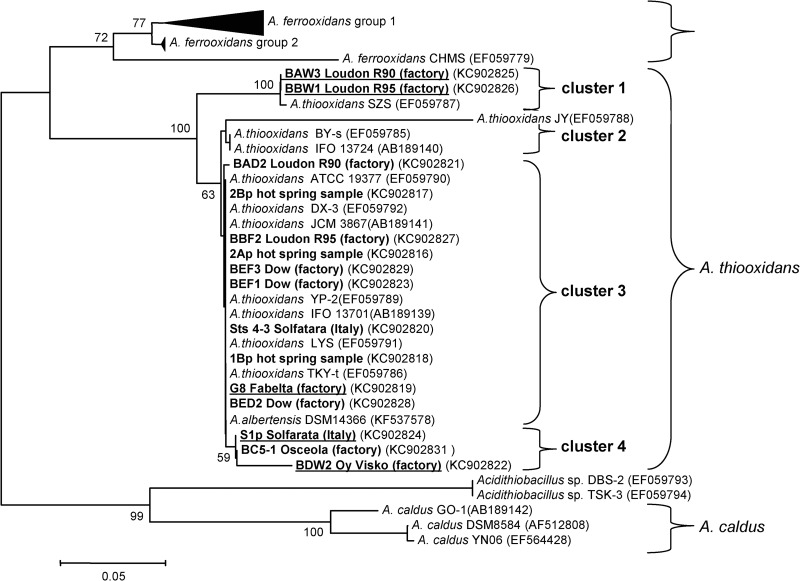
Fig 1.
Phylogenetic tree based on the 16S-23S ISR of CS2-converting A. thiooxidans strains isolated from environmental samples (1Bp, 2Ap, 2Bp, Sts 4-3, and S1p) or various industrial CS2 biofilter effluents (G8, BAD2, BED2, BBF2, BBW1, BAW3, BEF1, BDW2, BC6-1, and BEF3). The optimal neighbor-joining tree with a branch length sum of 0.98269283 is shown. The tree is drawn to scale, with branch lengths in the same units as the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed by the maximum composite likelihood method and are expressed as the number of base substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 330 positions in the final data set. The percentage above 50% of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. Four clusters of A. thiooxidans strains were identified, based on branch points that were reproduced in more than 50% of the bootstrap replicates. The type strains are A. thiooxidans ATCC 19377, A. albertensis DSM14366, and A. caldus DSM8584. In bold and underlined are strains that form white, large, dry, crusty colonies on Gelrite plates. In bold are strains that form small, shiny, smooth colonies on Gelrite plates.