Fig 1.
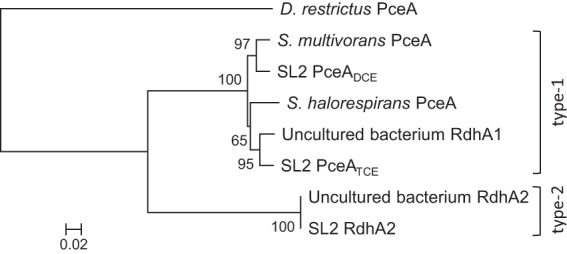
Sequence likelihood analysis showing the relationships of newly identified RdhA proteins from SL2 consortia to both Sulfurospirillum RdhA types. The neighbor-joining method of ClustalX was used to build the tree, including 100× bootstrap values. All sequences used in the alignment had similar lengths. The following sequences were used: S. multivorans PceA (GenBank accession no. AAC60788), S. halorespirans PceA (GenBank accession no. AAG46194), RdhA1 from an uncultured bacterium (GenBank accession no. BAJ09319), and RdhA2 from an uncultured bacterium (GenBank accession no. BAJ09321). The tree was rooted with PceA of Dehalobacter restrictus (GenBank accession no. CAD28790). The scale bar represents 2% sequence divergence.
