Abstract
The Gram-positive, anaerobic, spore-forming bacterium Clostridium perfringens causes a variety of diseases in both humans and animals, and spore germination is thought to be the first stage of C. perfringens infection. Previous studies have indicated that the germinant receptor (GR) proteins encoded by the bicistronic gerKA-gerKC operon as well as the proteins encoded by the gerKB and gerAA genes are required for normal germination of C. perfringens spores. We now report the individual role of these GR proteins by analyzing the germination of strains carrying mutations in gerKA, gerKC, or both gerKB and gerAA. Western blot analysis was also used to determine the location and numbers of GerKC proteins in spores. Conclusions from this work include the following: (i) gerKC mutant spores germinate extremely poorly with KCl, l-asparagine, a mixture of asparagine and KCl, or NaPi; (ii) gerKC spores germinate significantly more slowly than wild-type and other GR mutant spores with a 1:1 chelate of Ca2+ and dipicolinic acid and very slightly more slowly with dodecylamine; (iii) the germination defects in gerKC spores are largely restored by expressing the wild-type gerKA-gerKC operon in trans; (iv) GerKC is required for the spores' viability, almost certainly because of the gerKC spores' poor germination; and (v) GerKC is located in the spores' inner membrane, with ∼250 molecules/spore. Collectively, these results indicate that GerKC is the main GR protein required for nutrient and nonnutrient germination of spores of C. perfringens food-poisoning isolates.
INTRODUCTION
Clostridium perfringens is a Gram-positive, anaerobic, spore-forming pathogenic bacterium causing gastrointestinal (GI) diseases in humans and animals (1–3). The most important type that causes C. perfringens-associated food poisoning (FP) in humans is C. perfringens type A, and this illness is the third most commonly reported food-borne disease in the United States (1, 4). C. perfringens spores are resistant to many environmental stresses and remain dormant in the environment for a long period of time (5, 6). Once conditions are favorable, they can break their dormancy and initiate germination in response to a variety of compounds (called germinants), including amino acids, a mixture of a 1:1 chelate of Ca2+ and pyridine-2,6-dicarboxylic acid (dipicolinic acid [DPA]), and other nonnutrient compounds like dodecylamine, a cationic surfactant (7, 8).
In Bacillus subtilis spores, binding of specific nutrient germinants to their cognate germinant receptors (GRs) (7, 9–11) located in the spore's inner membrane triggers the following events: (i) release of monovalent cations, (ii) release of the spore core's large depot (∼20% of core dry weight) of DPA, and (iii) hydrolysis of the spore's peptidoglycan (PG) cortex by one or more spore cortex lytic enzymes that allows water uptake into the core to the level found in vegetative cells and then resumption of metabolism; spores also lose most of their resistance properties upon completion of germination (12, 13). B. subtilis spores have three major GRs, GerA, GerB, and GerK, each encoded by tricistronic operons that are expressed only in the developing forespore late in sporulation (10, 14). Loss of the A, B, or C cistron in these tricistronic GR operons leads to a loss of function of that GR (10, 15). Several studies (11, 16, 17) have shown that B. subtilis GerAA, GerAC, GerBA, and GerKA are located in the spore's inner membrane, with much of these proteins exposed on the outer surface of the inner membrane (18).
In C. perfringens, the organization of GR genes is different from that in B. subtilis (8, 19). There is no tricistronic gerA-like operon but rather a monocistronic gerAA located rather far from a gerK locus. The gerK locus includes a monocistronic gerKB in an orientation opposite of that of a bicistronic gerKA-gerKC (8). Previous work has shown that gerKA and/or gerKC is required for normal germination of C. perfringens spores with germinants such as KCl, l-asparagine, or an l-asparagine–KCl mixture termed AK (8). However, the gerAA and gerKB genes have at most only a minor role in normal spore germination with KCl (8, 19). In this study, we have evaluated the role of the GerKA and GerKC proteins in the germination of C. perfringens spores. The work has demonstrated that GerKC is the most important GR protein in the germination of C. perfringens spores and, further, that GerKC is located in the spores' inner membrane, with at least some of the protein being found on this membrane's outer surface.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The C. perfringens strains and plasmids used in this study are described in Fig. 1 and Table S1 in the supplemental material.
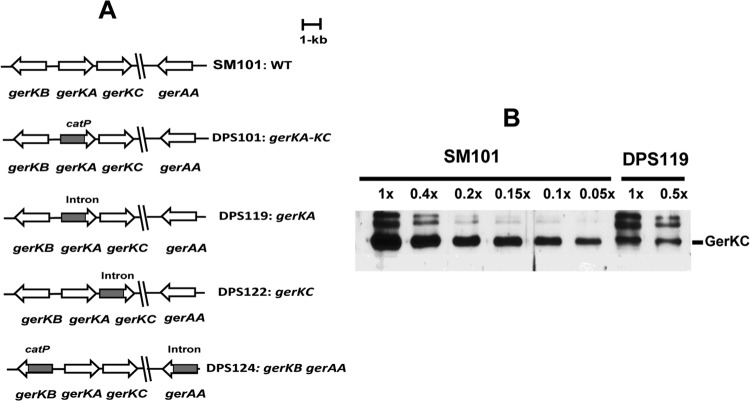
Fig 1.
C. perfringens GR mutants. (A) Arrangement of GR genes in various C. perfringens mutant strains. (B) Analysis of GerKC levels in total lysates of C. perfringens SM101 (wild-type [WT]) and DPS119 (ΔgerKA) spores. Spores were decoated and lysed, and various amounts of protein in the total lysates from the spores of the two strains were run on SDS-polyacrylamide gels, followed by Western blotting using GerKC antibody, as described in Materials and Methods. The intensity of the GerKC band in the DPS119 lysate was compared to the intensities of bands given by various amounts of GerKC in the SM101 lysate (1× corresponds to the same amount of spores, ∼1.5 × 107, in the two fractions). Analysis of the various band intensities by ImageJ gave a value for the GerKC level in DPS119 spores that was 12% of that in SM101 spores. The two bands above the GerKC band are likely bands that reacted nonspecifically, since their intensities were greatly increased relative to the intensity of the GerKC band in the DPS119 spores.
Construction of a gerKC mutant.
Construction of a gerKC mutant of C. perfringens strain SM101 was based on the modified group II intron (the ClosTron) using the ClosTron website to retarget a suitable insertion at bp 468/469 from the start codon of gerKC. PCR was performed on SM101 DNA to retarget the intron by using gerKC-specific primers CPP779, CPP780, and CPP781 (see Table S2 in the supplemental material) and the LtrBAsEBS2 universal primer (CGAAATTAGAAACTTGCGTTCAGTAAAC) provided with the Targetron gene knockout system (Sigma-Aldrich Corporation, St. Louis, MO). An ∼350-bp BsrGI-HindIII fragment retargeted to position 468/469 in the sense orientation of gerKC was cloned between the BsrGI and HindIII sites of the pJIR3566 vector (20), giving plasmid pDP276. Plasmid pDP276 was electroporated (21) into C. perfringens strain SM101, and erythromycin (Em)-resistant (Emr) transformants were selected on brain heart infusion (BHI) agar plates supplemented with 30 μg/ml Em. Emr transformants were screened for insertion of the Targetron by PCR using gerKC-specific primers CPP440 and CPP443 (see Table S2 in the supplemental material). To cure the Emr-coding vector, one candidate Targetron-carrying clone was subcultured three times in nonselective fluid thioglycolate (FTG) broth and plated onto BHI agar, and single colonies were patched onto BHI agar with or without Em, giving strain DPS122.
Construction of a gerKA mutant.
A C. perfringens SM101 derivative with an intron inserted into the gerKA gene was constructed as follows. To target the L1.LtrB intron to gerKA, the intron sequence in plasmid pJIR3566 was modified using the InGex intron prediction program (Sigma-Aldrich), and an insertion site at bp 90/91 (score, 10.5) from the start codon was chosen. PCR was performed on SM101 DNA to retarget the intron using gerKA-specific primers CPP776, CPP777, and CPP778 (see Table S2 in the supplemental material) and the LtrBAsEBS2 universal primer (CGAAATTAGAAACTTGCGTTCAGTAAAC) provided with the Targetron gene knockout system (Sigma-Aldrich). An ∼350-bp BsrGI-HindIII fragment was retargeted to gerKA and was cloned between the BsrGI and HindIII sites in pJIR3566, giving plasmid pDP300. Plasmid pDP300 was electroporated into C. perfringens strain SM101, and the gerKA mutant strain was isolated essentially as described above for the gerKC mutant.
Construction of a gerKB gerAA double mutant.
A mutation in the gerAA gene of a previously isolated gerKB mutant strain, DPS108 (gerKB::catP) (19), was generated by inserting a Targetron in the antisense strand of gerAA between bp 123 and 124 downstream of the gerAA start codon, as previously described (8). Briefly, the gerAA Targetron containing plasmid pDP13 carrying the Targetron insertion between bp 123 and 124 was introduced into DPS108 by electroporation, and clones were identified with an insertion of the intron into gerAA, giving a C. perfringens gerKB::catP gerAA::intron double mutant strain, DPS124.
Construction of a gerKC strain complemented with wild-type gerKA-gerKC.
To construct a gerKC strain complemented with wild-type gerKA-gerKC, we first constructed a shuttle plasmid carrying the wild-type gerKA-gerKC operon. An ∼3.2-kb KpnI-SalI fragment carrying the gerKA-gerKC operon with its own promoter and terminator was excised from plasmid pDP10 (8) by digestion with KpnI and SalI and then ligated between the KpnI and SalI sites of the Escherichia coli-C. perfringens shuttle plasmid pJIR750 that encodes chloramphenicol resistance (Cmr) (22), giving plasmid pSB18. The recombinant plasmid pSB18 was introduced into C. perfringens gerKC mutant strain DPS122 by electroporation (21) and Cmr Emr transformants were selected. The presence of plasmid pSB18 in strain DPS122(pSB18) was confirmed by PCR and Southern blot analyses (data not shown).
Spore preparation.
Starter C. perfringens cultures were prepared by inoculating 0.1 ml of cooked meat stock culture into 10 ml FTG broth (Difco) and incubating at 37°C, as described previously (23). Overnight FTG-grown C. perfringens cultures (0.4 ml) were inoculated into 10 ml fresh FTG broth and incubated at 37°C for 8 to 12 h. To prepare sporulating cultures, 0.2 ml of FTG-grown culture was transferred into 10 ml Duncan-Strong broth (24) and incubated at 37°C for 24 h. Spore formation was confirmed by phase-contrast microscopy. Scaling up of this procedure was needed for large-scale spore preparation. For purification, spore suspensions were washed with sterile distilled water until they were >98% free of vegetative cells, germinated spores, and debris. Purified spores were resuspended in sterile distilled water at an optical density at 600 nm (OD600) of ∼6 and stored at −80°C.
Spore germination.
C. perfringens spore germination was done as previously described (8, 25). Briefly, heat-activated (heated at 80°C for 10 min and then cooled on ice) spore suspensions (OD600, ∼1) were incubated with KCl (100 mM KCl in 25 mM Tris-HCl buffer, pH 7.0), l-asparagine (100 mM l-asparagine in 25 mM Tris-HCl buffer, pH 7.0), l-asparagine plus KCl (100 mM l-asparagine–100 mM KCl in 25 mM Tris-HCl buffer, pH 7.0), NaPO4 buffer (100 mM NaPO4 buffer, pH 6.0), or Ca-DPA (50 mM CaCl2, 50 mM DPA, adjusted to pH 8.0 with Tris-HCl) at 40°C, and spore germination was measured by monitoring the OD600 of germinating spores in a Synergy Mx multimode microplate reader (BioTek Instruments Inc., Winooski, VT). Levels of spore germination were also routinely confirmed at the end of the experiments by phase-contrast microscopy. The extent of spore germination was expressed as a percentage of the initial OD600, with a decrease of 60% seen with ∼100% spore germination. All values reported are averages of three experiments performed with at least three independent spore preparations.
For measuring DPA release during KCl-triggered germination, heat-activated spores (OD600, 1.5) were incubated at 40°C in 100 mM KCl in 25 mM Tris-HCl buffer (pH 7.0). DPA release during dodecylamine-triggered germination was measured by incubating heat-activated spore suspensions (OD600, 1.5) in 1 mM dodecylamine in 25 mM Tris-HCl (pH 7.4) at 60°C. Aliquots (1 ml) of spores whose germination was triggered by KCl or dodecylamine were centrifuged for 3 min at 13,000 rpm in a microcentrifuge, and the DPA in the supernatant was measured by monitoring the OD at 270 nm as previously described (8, 26, 27). Initial DPA levels in dormant spores were measured by boiling 1-ml aliquots for 60 min and centrifugation for 3 min at 13,000 rpm, and the DPA content of the supernatant was measured at 270 nm (8, 26, 27).
Preparation of the spore IM fraction.
The spore inner membrane (IM) was prepared as described previously (16, 18, 28–30). Spores (∼3 ml) at an OD600 of 25 were first decoated by incubation at 70°C for 2 h in 0.1 M NaOH, 0.1 M NaCl, 0.5% SDS, 0.1 M dithiothreitol (DTT) and washed 10 times with water, as described previously (31). The washed decoated spores of wild-type strain SM101 and strain DPS122 were suspended at an OD600 of 50 to 70 in 0.5 ml TEP buffer (50 mM Tris-HCl [pH 7.4], 5 mM EDTA, 1 mM phenylmethanesulfonyl fluoride [PMSF]) with 1 mg lysozyme, 1 μg each of RNase A and DNase I, and 20 μg MgCl2, as described previously (16, 18, 29, 32). The spore suspension was then incubated for 5 min at 37°C. After cooling on ice for 20 min, suspensions were briefly sonicated with 100 mg glass beads, examined microscopically for lysis, and centrifuged for 5 min in a microcentrifuge at maximum speed, and the supernatant was saved. The pellet was resuspended in 0.5 ml TEP buffer and centrifuged again as described above, the supernatants were pooled, and the pellet was also saved. The pooled supernatant was centrifuged at 100,000 × g for 1 h at 4°C in an ultracentrifuge with a TLS 100.2 rotor, giving a soluble fraction (S100) and the IM pellet fraction (P100), which was suspended in 40 to 80 μl of TEP buffer containing 1% Triton X-100.
GerKC expression and purification and antibody production.
The gerKC gene was amplified by PCR using genomic DNA from C. perfringens strain SM101 as the template and cloned into a modified pET15b expression vector containing a tobacco etch virus (TEV) protease cleavage site between the N-terminal His6 tag and the target gene. The identity of the resultant plasmid was verified by DNA sequencing. The GerKC protein (residues 28 to 374) was overexpressed in Escherichia coli BL21 cells and purified by Ni2+-nitrilotriacetic acid affinity chromatography, followed by TEV protease cleavage of the His6 tag and by anion exchange and gel filtration chromatography (SD200; GE Healthcare, Piscataway, NJ). Purified protein (∼2 mg) was used to raise polyclonal antibodies in two New Zealand White female rabbits (Pacific Immunology Group, Ramona, CA). Anti-GerKC antibodies were detected by Western blotting in a blood sample collected 2 months after the initial injection, at which time the animals were exsanguinated. The specificity of the antiserum was confirmed by Western blotting using purified GerKC protein (see Results).
Animals were used by Pacific Immunology only to prepare antibody in their facility (NIH Animal Welfare Assurance number A41820-01; USDA license 93-R-283). Their facility adheres to the standards and regulations under the Animal Welfare Act and requires that any exceptions to the Act be approved by the IACUC.
Western blot analysis.
Western blot analyses were carried out following SDS-polyacrylamide (12%) gel electrophoresis (SDS-PAGE) and transfer of proteins to polyvinylidene difluoride (PVDF) membranes as described previously (13, 18, 32). The GerKC protein was detected by using polyclonal rabbit antisera (1:1,000) against GerKC and goat anti-rabbit IgG–horseradish peroxidase (HRP) conjugate (1:10,000). The activity of HRP was detected using a PicoMax sensitive chemiluminescent HRP substrate (Rockland Immunochemicals, Gilbertsville, PA) with a chemiluminescence detection system (Molecular imagDoc XRS+ system; Bio-Rad, Hercules, CA).
Determination of GerKC level in spores.
Dormant spores (∼20 mg) of C. perfringens strain SM101 (wild type) were decoated, treated with lysozyme, disrupted with glass beads and sonication (∼8.4 × 109 spores were disrupted), and then centrifuged at 14,000 × g for 10 min as described above. The pellet from this initial centrifugation was saved and termed the pellet fraction. The supernatant fraction was further centrifuged as described above, giving inner membrane pellet and supernatant fractions, and various percentages of the inner membrane and initial pellet fractions along with known quantities of purified GerKC were subjected to Western blot analysis using anti-GerKC serum. The intensities of the resulting bands were quantitated using ImageJ software, and the average total number of GerKC molecules per spore was calculated (29, 32).
Determination of GerKC susceptibility to modification by an exogenous biotinylation reagent.
Biotinylation was performed as previously described (18) with some modifications. Dormant SM101 spores at an OD600 of ∼25 (5 ml) were decoated and washed as described above. Buoyant density gradient centrifugation was performed in 50% Histodenz medium (Sigma Chemical Co., St. Louis, MO) to remove small amounts of germinated spores. The dormant spore pellet from this centrifugation was washed extensively with water, and the resulting decoated dormant spores were suspended in water at an OD600 of 100.
Germinated spores were obtained by first heat shocking intact dormant spores (3 ml at an OD600 of 50) at 80°C for 10 min. After cooling to 25°C for 5 min, spores were heated to 40°C for 10 min and germinated at an OD600 of ∼1.5 in 50 ml of 100 mM KCl in 25 mM sodium phosphate buffer (pH 7.0) for 2 h, until at least 90% of spores were germinated, as determined by phase-contrast microscopy. Small amounts of dormant spores were not removed from these germinated spore preparations. However, these dormant spores had not been decoated and were thus not lysed by subsequent lysozyme treatment. Consequently, the IM was not isolated from these dormant spores but was isolated only from the spores that had become lysozyme sensitive due to their germination.
Biotinylation of lysyl amino groups in dormant or germinated spore protein was done with 5 ml of spores at an OD600 of ∼15 in 10 mM K-HEPES buffer (pH 7.4) and 150 mM NaCl using 2 mM EZ-Link sulfo-NHS-SS-biotin reagent (Pierce Chemical Co., Rockford, IL) for 1 h at room temperature on a rocker. Unreacted reagent was quenched by addition of 1 ml of 2 M glycine and 1 ml of 1 M Tris-HCl buffer (pH 7.4) and incubation for 30 min at 23°C. All labeled spores were washed twice with 20 ml water.
For analysis of GerKC biotinylation, NeutrAvidin agarose beads (Pierce) were prepared by spinning 100 μl of slurry at 1,000 × g for 1 min at 23°C, discarding the supernatant, and then washing twice with 100 μl TEP buffer containing 1% Triton X-100. Sixty microliters of the IM fraction was added to the resulting bead pellet, and the mixture was shaken on a rocker. After shaking at 23°C for 1 h, the sample was centrifuged at 1,000 × g for 1 min and the supernatant containing unbound proteins () was saved. The resulting bead pellet was washed three times with 100 μl TEP buffer containing 1% Triton X-100, and the washes were also saved. Bound proteins were eluted by incubating the beads with 60 μl Laemmli sample buffer containing 55 mM DTT for 1 h at 23°C. Western blot analyses were performed as described above following SDS-PAGE of equal percentages of the total IM fraction, the flowthrough fraction, and the eluted fraction, all run on the same Western blot and in some cases with various dilutions of the flowthrough or eluted fraction.
Other methods.
For determination of the colony-forming efficiencies of spores of various strains, spores at an OD600 of 1 (∼108 spores/ml) were heat activated as described above, aliquots of dilutions were spread on BHI agar with or without lysozyme (1 μg/ml) and incubated anaerobically at 37°C for 24 h, and colonies were counted.
Student's t test was used for specific comparisons between data sets.
RESULTS
gerKA mutant spores produce lower levels of GerKC than wild-type spores.
Previous studies (8, 19) suggested that the main GR proteins involved in the germination of C. perfringens spores are encoded by the bicistronic gerKA-gerKC operon, while the products of gerKB and gerAA have auxiliary roles. However, those studies were conducted on a C. perfringens strain (DPS101) with an intron insertion into the gerKA cistron that likely had a polar effect on expression of the downstream gerKC. To dissect the role of the GerKA and GerKC GR proteins individually in C. perfringens spore germination, we constructed mutants with double gerAA gerKB mutations and single gerKA or gerKC mutations in strain SM101 (Fig. 1A). To evaluate whether insertion of an intron in gerKA exerted a polar effect on the downstream gerKC, the levels of GerKC in spores of wild-type and gerKA mutant strains were compared (Fig. 1B). Western blotting with antisera against GerKC demonstrated the presence of GerKC in gerKA mutant spores. However, quantitative analyses showed that there was only ∼12% of the wild-type GerKC level in gerKA mutant spores. These results indicate that the gerKA mutation did indeed exert a partial polar effect on the downstream gerKC gene.
GerKC, but not GerKA, has a major role in C. perfringens spore germination.
Spores of the double gerAA gerKB mutant strain germinated well in the presence of l-Asn, KCl, AK, or NaPi (Fig. 2A to D), confirming the minor roles played by GerKB and GerAA in spore germination (8, 19) and consistent with GerKA and/or GerKC being the main GR proteins in spores of C. perfringens FP strain SM101.
Fig 2.
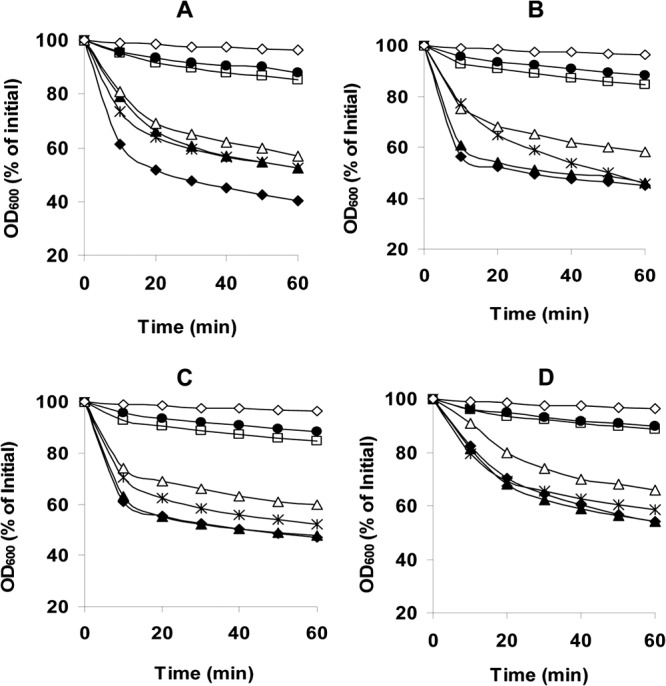
Germination of C. perfringens spores with various germinants. Heat-activated spores of strains SM101 (wild type) (◆), DPS101 (gerKA-KC) (●), DPS119 (gerKA) (▲), DPS122 (gerKC) (□), DPS124 (gerKB gerAA) (×), and DPS122(pSB18) (gerKC mutant complemented with wild-type gerKA-gerKC) (△) were incubated at 40°C with KCl (A), l-Asn (B), AK (C), or NaPi (pH 6.0) (D), and germination was measured by determination of the OD600, as described in Materials and Methods. The control germination (♢) was heat activation of spores, which were incubated in 25 mM Tris-HCl buffer (pH 7.0) at 40°C, and no germination difference was observed between spores of SM101 and the GR mutant strains.
To dissect the precise roles of GerKA and GerKC in C. perfringens spore germination, we constructed a single gerKA or gerKC mutant in strain SM101 (Fig. 1) and measured the germination of the spores of these various strains. As expected, gerKA gerKC spores germinated poorly and to a much lesser extent than wild-type spores with l-Asn, KCl, AK, or NaPi (Fig. 2A to D). While gerKA and wild-type spores germinated similarly, gerKC spores germinated much more poorly with these germinants than wild-type and other GR mutant spores (P < 0.05) (Fig. 2A to D). Most importantly, the germination defects of gerKC spores were largely corrected by complementing the gerKC mutant with the wild-type gerKA-gerKC operon. Collectively, these results indicate that GerKC is the most important GR protein in C. perfringens SM101 spore germination.
After germinants bind their cognate GR, the next easily measurable event in spore germination is the release of DPA from the spore core. Therefore, we also evaluated the effects of mutations in GR protein genes on DPA release during germination with KCl (Fig. 3). As expected, wild-type spores released ∼70% of their DPA within the first 10 min of germination with KCl (Fig. 3). Strikingly, the rate and amount of DPA release by gerKA spores incubated with KCl were similar to those of wild-type spores, indicating that GerKA is not essential for DPA release during spore germination, at least when GerKB and GerAA are present, and only slightly less DPA release was obtained when gerAA gerKB spores were germinated with KCl. In contrast, gerKC spores released ∼50% less DPA (P < 0.05) than wild-type spores during KCl germination (Fig. 3). The DPA release defect in gerKC spores was also corrected by complementation with a plasmid carrying wild-type gerKA-gerKC. These results further indicate that GerKC is the main GR protein involved in C. perfringens spore germination.
Fig 3.
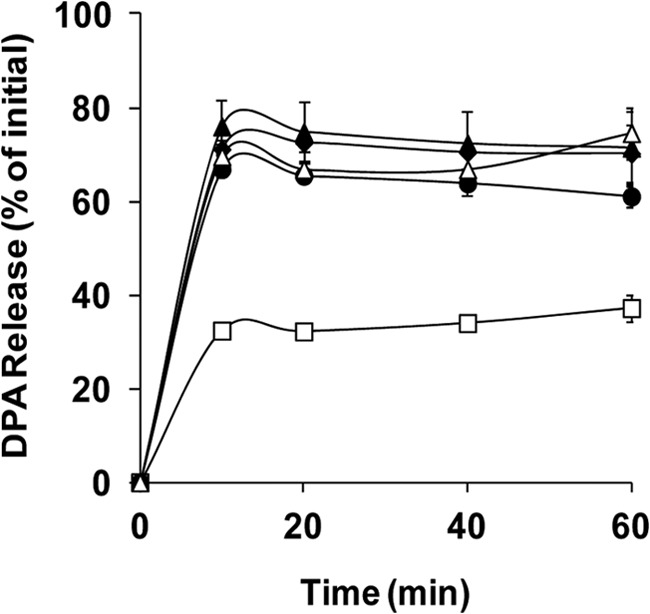
DPA release by spores of C. perfringens strains during germination with KCl. Heat-activated spores of strains SM101 (wild-type) (◆), DPS119 (gerKA) (▲), DPS122 (gerKC) (□), DPS124 (gerKB gerAA) (●), and DPS122(pSB18) (gerKC mutant complemented with wild-type gerKA-gerKC) (△) were germinated with KCl (pH 7.0), and DPA release was measured as described in Materials and Methods. Error bars represent standard deviations.
GerKC is also important in germination of C. perfringens spores with Ca-DPA and dodecylamine.
Germination of bacterial spores can also be triggered by nonnutrient germinants, such as the cationic surfactant dodecylamine (27) and exogenous Ca-DPA, and in B. subtilis spores, germination with these agents does not involve any GR protein (7, 15). It was previously shown that unlike spores of Bacillus species, where Ca-DPA triggers spore germination by direct activation of the cortex-lytic enzyme CwlJ (9, 33), in C. perfringens, Ca-DPA triggers germination in a GerKA- and/or GerKC-dependent pathway (8). When wild-type, gerKB gerAA, and gerKA spores were incubated with Ca-DPA, significant germination was observed, as measured by the decrease in the OD600 (Fig. 4A). In contrast, there was minimal germination of gerKC spores with Ca-DPA (Fig. 4A), and phase-contrast microscopy confirmed that ∼95% of these spores remained phase bright (data not shown). The Ca-DPA germination defect of the gerKC spores was partially corrected by complementation with wild-type gerKA-gerKC (Fig. 4A), indicating that the Ca-DPA germination pathway is dependent on GerKC.
Fig 4.
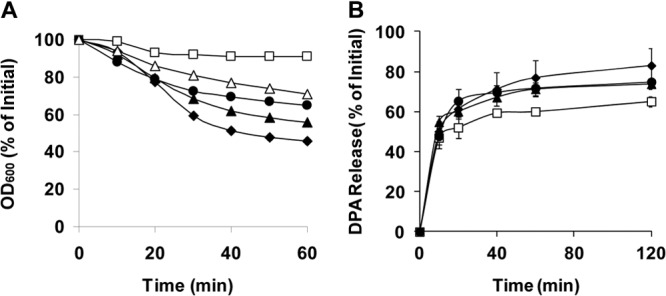
Germination of C. perfringens wild-type and mutant spores with Ca-DPA (A) and dodecylamine (B). Heat-activated spores of strains SM101 (wild-type) (◆), DPS119 (gerKA) (▲), DPS122 (gerKC) (□), DPS124 (gerKB gerAA) (●), and DPS122(pSB18) (gerKC mutant complemented with wild-type gerKA-gerKC) (△) were germinated with Ca-DPA and dodecylamine. For Ca-DPA, changes in OD600 were measured as described in Materials and Methods, and for dodecylamine, germination was monitored by measurement of DPA release, as described in Materials and Methods. Error bars represent standard deviations.
Previous work showed that GerKA and/or GerKC is also required for normal spore germination with dodecylamine (8). In the current work, gerKB gerAA and gerKA spores released almost the same amount of DPA as wild-type spores during germination with dodecylamine (Fig. 4B), suggesting that GerAA, GerKB, and GerKA might have no role in dodecylamine germination. The gerKC spores incubated with dodecylamine released slightly less DPA than wild-type spores (Fig. 4B), indicating that GerKC may play a minor role in triggering DPA release during dodecylamine germination.
GerKC is required for normal colony formation in BHI medium.
Unlike B. subtilis, where spores lacking all GRs retain full viability once these are decoated and rescued in the presence of lysozyme, the absence of gerKA-gerKC and gerKB does affect the apparent viability of C. perfringens spores (8, 19). Therefore, to establish if it is GerKA or GerKC that is involved in C. perfringens spores' apparent viability, the colony-forming efficiencies of intact and decoated wild-type and mutant C. perfringens spores were assessed by plating spores onto BHI agar and incubating them anaerobically for 24 h at 37°C. There was no significant (P >0.05) difference in the colony formation efficiency of wild-type and gerKA spores, while gerAA gerKB spores had an ∼6-fold lower colony formation efficiency (Table 1). However, intact gerKC spores exhibited an ∼100-fold lower colony formation efficiency than wild-type and gerKA spores (Table 1). Decoating of spores and plating on BHI agar containing lysozyme increased the colony-forming efficiencies of spores of all mutant strains tested, including the gerKC spores, to a level approximately equal to that of the wild-type spores (Table 1). Thus, the gerKC spores are fully viable, and most simply do not germinate well.
Table 1.
Colony formation by spores of C. perfringens strainsa
| Strain (genotype) | Spore titer (no. of CFU/ml/OD600 unit)b |
|
|---|---|---|
| BHI | BHI + Lyzc | |
| SM101 (wild type) | 2.4 × 108 | 7.1 × 108 |
| DPS 119 (gerKA) | 1.7 × 108 | 5.7 × 108 |
| DPS122 (gerKC) | 2.3 × 106 | 4.2 × 108 |
| DPS124 (gerKB gerAA) | 3.8 × 107 | 6.1 × 108 |
| DPS122(pSB18) (gerKC mutant complemented with wild-type gerKA-gerKC) | 1.2 × 108 | NDd |
Heat-activated spores of various strains were plated on BHI agar with or without lysozyme, and colonies were counted after anaerobic incubation at 37°C for 24 h, as described in Materials and Methods.
Titers are the average number of CFU/ml/OD600 unit determined in three experiments, and the variation was less than 15%.
Spores were decoated, heat activated, and plated onto BHI agar containing lysozyme (Lyz), and colonies were counted after they were incubated overnight anaerobically at 37°C.
ND, not determined.
To directly examine the possibility that the lower colony formation efficiency of gerKC spores was due to their poor germination in BHI broth, we compared the germination of spores of all four strains in this medium. As expected, gerKC spores exhibited significantly (P < 0.05) lower germination in BHI broth than wild-type spores, and the germination defect in gerKC spores was at least partially restored by complementing the mutant with wild-type gerKA-gerKC (Fig. 5). The gerKA and gerKB gerAA spores also exhibited slightly lower germination than wild-type spores (Fig. 5). Phase-contrast microscopy found that >65% of wild-type and gerKA spores, ∼50% of gerKB gerAA and gerKC-complemented spores, but only ∼10% of gerKC spores were phase dark after 1 h of incubation in BHI broth (data not shown), in agreement with the results obtained by measurements of germination by determination of the OD600 (Fig. 5). In addition, when the spores were incubated aerobically for 18 h in BHI broth, ∼95% of wild-type, gerKA, gerKB gerAA, and gerKC-complemented spores were phase dark, and ∼90% of these phase-dark spores had released the nascent vegetative cell (data not shown). In contrast, only ∼65% of gerKC spores were phase dark and less than 10% of the phase-dark spores had released the nascent vegetative cell (data not shown), again consistent with the lower colony formation efficiency observed with these spores on plates without lysozyme. Collectively, these results indicate that GerKC, but not GerKA, is essential for normal completion of spore germination and perhaps also outgrowth and, thus, for normal colony formation in BHI medium.
Fig 5.
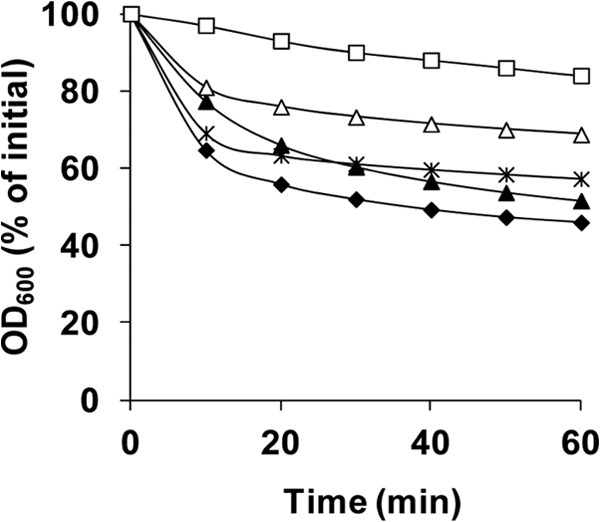
Germination of spores of C. perfringens strains in BHI broth. Heat-activated spores of strains SM101 (wild-type) (◆), DPS119 (gerKA) (▲), DPS122 (gerKC) (□), DPS124 (gerKB gerAA) (×), and DPS122(pSB18) (gerKC mutant complemented with wild-type gerKA-gerKC) (△) were germinated at 40°C in BHI broth, and the OD600 was measured as described in Materials and Methods.
GerKC localizes to the IM of C. perfringens spores.
The GR proteins of Bacillus species have been localized in spores' IM (11, 16, 17). Therefore, since GerKC is essential for germination of C. perfringens spores, we attempted to immunolocalize GerKC in spores using polyclonal rabbit antiserum raised against recombinant C. perfringens GerKC, and the recombinant GerKC was detected as an ∼40-kDa protein by the antiserum (Fig. 6). When spore IM fractions obtained from decoated C. perfringens wild-type and gerKC spores were separated by SDS-PAGE and subjected to Western blot analysis (Fig. 6), a dominant 40-kDa protein band was detected in the IM fraction from wild-type but not gerKC spores. These results clearly indicate that the GerKC localizes to the IM of C. perfringens spore.
Fig 6.
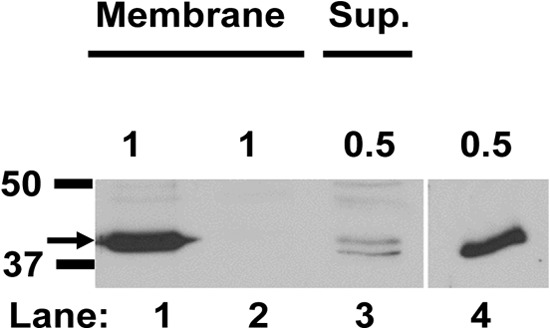
Localization of GerKC in C. perfringens spores. Dormant spores of C. perfringens strains SM101 (wild type) and DPS122 (gerKC) were decoated, disrupted with glass beads and sonication after lysozyme treatment, and centrifuged at 14,000 × g for 10 min, and the supernatant was centrifuged at 100,000 × g for 1 h, as described in Materials and Methods. Aliquots of the P100 pellet IM fractions (lanes 1 and 2) from wild-type and gerKC spores and the S100 supernatant (Sup.) fraction from wild-type spores (lane 3) were run on SDA-polyacrylamide gels, followed by Western blotting using GerKC antibody, as described in Materials and Methods. The arrow on the left indicates the size of GerKC, ∼40 kDa. The amounts of sample run in lanes 1 to 3 are given above the lanes as the percentage of the total IM protein or supernatant fraction protein. The S100 supernatant fraction from gerKC spores had no detectable GerKC band (data not shown). Lane 4, 0.5 ng purified GerKC antigen. The numbered lines to the left of the figure denote the migration positions of molecular mass markers (in kDa). All lanes are from the same gel, but an intervening region between lanes 3 and 4 was removed for clarity.
C. perfringens has ∼250 GerKC molecules per spore.
In bacterial spores, GRs act as environmental sensors that trigger spore germination when conditions are deemed adequate for growth (7, 10, 34). The germination machinery in B. subtilis spores appears to amplify the germination signal through increases in the levels of various germination components, with the GRs being present in hundreds of molecules per spore, and with the proteins involved downstream of GRs, the SpoVA proteins that are essential for DPA release in spore germination, being present at 5,000 to 10,000 molecules per spore (16, 17, 28, 32, 35). Consequently, it was of interest to analyze the level of GerKC in C. perfringens spores. In order to obtain a complete assessment of GerKC levels in spores, the IM fraction and the pellet fraction obtained by low-speed centrifugation after spore disruption were analyzed for GerKC by Western blotting, and the intensities of the GerKC bands in these fractions were compared to those of purified GerKC (Fig. 7). While ∼60% of the GerKC was present in the IM fraction, ∼40% was found in the pellet fraction obtained by low-speed centrifugation, presumably reflecting the presence of significant amounts of inner membrane in this fraction as well. The same results were also found for spores of B. subtilis (32). In total, C. perfringens spores were calculated to have ∼250 GerKC molecules per spore, although this number may vary significantly between individual spores in the population.
Fig 7.

Quantitation of the GerKC level in spores of C. perfringens strain SM101. Various amounts of P100 pellet IM fractions (lanes 1 to 3) and pellet fractions of decoated, disrupted spores obtained by centrifugation at 14,000 × g (lanes 4 and 5) and various amounts of purified GerKC antigen (lanes 6 to 8) were run on SDS-polyacrylamide gels, followed by Western blot analysis using anti-GerKC antibody. The amounts of samples run are shown above the lanes as either the percentage of the total membrane protein or pellet fraction protein from 8.4 × 109 spores (lanes 1 to 5) or the amount (in ng) of GerKC antigen (lanes 6 to 8). The numbered lines to the left of the figure denote the migration positions of molecular mass markers (in kDa). All lanes are from the same gel, but lanes 4 and 5 were moved from their original position to the right of the GerKC antigen lanes.
Topology of GerKC in C. perfringens spores' IM.
Analysis of the amino acid sequence encoded by the gerKC gene indicates that GerKC is likely a peripheral membrane lipoprotein. While most of the protein is relatively hydrophilic, it has a hydrophobic N-terminal region that is likely a signal peptide that is followed by a consensus sequence for covalent addition of a diacylglycerol moiety to an N-terminal cysteine residue formed by signal peptide cleavage. A high-resolution structure of a B. subtilis GR C protein has been determined and is a relatively globular structure (36). Even more importantly, prediction of the structure of other GR C proteins, including those from Clostridium species, suggests that all GR C proteins adopt a similar structure. Given this likely structure of C. perfringens GerKC, as well as the expression of gerKA-gerKC only in the developing forespore, it is thus most likely that GerKC is a peripheral membrane protein on the outer leaflet of the spore IM.
Analysis of the topology of GR C proteins by analysis of these proteins' susceptibility to biotinylation by an exogenous reagent has recently shown that these proteins are well biotinylated in intact B. subtilis spores, suggesting that GR C proteins are indeed on the IM outer surface (18). Consequently, we also subjected intact dormant and germinated C. perfringens spores to the same analysis and evaluated GerKC for biotinylation by Western blotting following affinity purification of biotinylated proteins (Fig. 8). Approximately 33% of C. perfringens GerKC was biotinylated in decoated but otherwise intact C. perfringens spores, with this value increasing to ∼50% in intact germinated spores. This value for dormant spores is almost identical to that found for the biotinylation of two GR C proteins in dormant B. subtilis spores, although the same two GR C proteins were 74 to 100% biotinylated in germinated B. subtilis spores (18). These results are certainly consistent with the majority of GerKC being located on the outer surface of C. perfringens spores' IM, although we cannot be sure that all GerKC is there. We also do not understand why biotinylation of GerKC in dormant and germinated spores is not complete, although this has also been seen with several B. subtilis IM proteins, including GR subunits (18).
Fig 8.
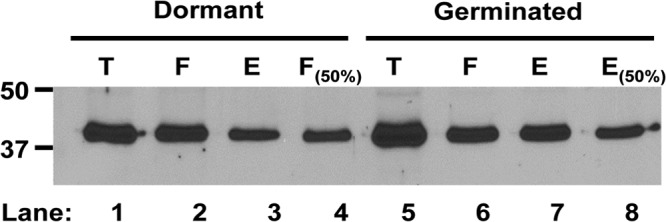
Analysis of the level of biotinylation of GerKC in decoated dormant and germinated spores of C. perfringens wild-type strain SM101. Biotinylation was carried out on decoated dormant spores and intact germinated spores as described in Materials and Methods. Spores were disrupted and fractionated, and the levels of various germination proteins in (i) the total IM fraction (T), (ii) the IM fraction that did not adsorb to the NeutrAvidin beads (F), and (iii) the eluate from the NeutrAvidin beads (E) were assayed by Western blotting using anti-GerKC antiserum, as described in Materials and Methods. Aliquots in the T, F, and E samples were from equal amounts of spores. The F(50%) sample (lane 4) from dormant spores was also the protein that did not adsorb to the NeutrAvidin beads but was from 1/2 the amount of spores as in the other lanes. The E(50%) sample (lane 8) from germinated spores was also the eluate from the NeutrAvidin beads but was from 1/2 the amount of spores as in the other lanes. Analysis of this Western blot by the program ImageJ indicated that ∼33% of GerKC was biotinylated in dormant spores, with ∼50% biotinylated in germinated spores.
DISCUSSION
A first major conclusion from this work is that GerKC is the major GR protein in spores of C. perfringens food-poisoning strain SM101. Previous work (8) suggested that the products of the bicistronic gerKA-gerKC operon were most important in spore germination; however, those studies were done on a C. perfringens strain with a mutation on the gerKA cistron, which caused a polar effect on the downstream gerKC. Here, we show that a Targetron mutant with a mutation in gerKA still allows significant germination of gerKA spores, suggesting that GerKC plays a major role in this process. In fact, inactivation of gerKC in strain SM101 affected spore germination with all known nutrient germinants. However, it is unclear whether GerKC acts as a receptor itself and contains multiple ligand binding sites, is an essential component of a multiple-subunit receptor formed by GerAA, GerKB, and/or GerKA, or even interacts with other unknown proteins. Clearly, further studies are needed to answer these questions.
Another major conclusion is that GerKC is required for full apparent viability of C. perfringens spores. The lower colony-forming ability of gerKC spores in rich BHI medium compared to that of wild-type and gerKA spores was consistent with the significantly slower germination of gerKC spores in BHI medium and the slower release of nascent vegetative cells than wild-type and gerKA spores. The reversion to wild-type levels of colony formation by gerKC spores complemented with wild-type gerKA-gerKC is further evidence that GerKC is also important in spore viability. However, the lower viability of gerKC spores is apparent only since full gerKC spore viability was restored when decoated spores were plated in the presence of lysozyme. Perhaps the GerKC protein is involved in the activation of either the cortex lytic enzyme (CLE) SleC to allow cortex hydrolysis or some unknown enzyme that induces the release of nascent vegetative cells from the coat/exosporium. This role of C. perfringens GerKC is in contrast to that of B. subtilis GRs, where the viability of spores lacking one GR is relatively normal, although spores lacking all GRs do exhibit low apparent viability (15).
A third major conclusion of this work is that GerKC localizes to the inner membrane of C. perfringens spores. This finding was not unexpected, since GR proteins of B. subtilis have been shown to localize in the spore inner membrane (11, 16, 17). The level of GerKC in C. perfringens spores was also just 2- to 4-fold lower than the levels of GR in B. subtilis spores. A key difference in the components of the germination machinery between C. perfringens and B. subtilis spores is that C. perfringens spores lack a homologue of the lipoprotein GerD (34), which plays a major role in GR-dependent germination in B. subtilis spores (30, 37), perhaps by ensuring that all GRs localize into the germinosome (32).
A minor conclusion from this work is that GerKC is essential for normal DPA release during C. perfringens spore germination with many germinants. Previous work (8) suggested that GerKA and/or GerKC is required for normal DPA release with nutrient and nonnutrient germinants. Here, we show that only the absence of GerKC affects DPA release from the spore core during nutrient and nonnutrient germination of C. perfringens spores. Further work will be needed to establish the reason for the apparent partial release of DPA during germination of gerKC C. perfringens spores. Some possible explanations for these results are that GerKC (i) is required for the correct colocalization or stability of other GR proteins and/or to form a functional GR by itself, with other GR subunits, or with unknown proteins that will efficiently trigger Ca-DPA release; (ii) might be acting as a link between GR subunits (i.e., GerAA, GerKB, and GerKA) and proteins that form the Ca-DPA channel; or (iii) is required for complete DPA release from individual spores during germination, although this seems unlikely, since DPA release from individual germinating spores of both C. perfringens and Bacillus species is invariably an all-or-none phenomenon (38–40).
In conclusion, the results presented in this work should provide a deeper insight into the mechanism of C. perfringens spore germination in which KCl, l-Asn, AK, NaPi, and the nonnutrient germinants Ca-DPA and dodecylamine act largely or at least in part through the GerKC GR protein. In contrast, GerKA, GerAA, and GerKB play at most auxiliary roles, since deletion of either of these GR proteins has no significant effect on C. perfringens spore germination.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the N. L. Tartar Foundation of Oregon State University, Agricultural Research Foundation of Oregon State University (to M.R.S), by a U.S. Department of Defense Multidisciplinary University Research Initiative (MURI) award through the U.S. Army Research Laboratory and the U.S. Army Research Office under contract number W911NF-09-1-0286 (to M.R.S., P.S., B.H.), and by grants from the Fondo Nacional de Ciencia y Tecnología de Chile (FONDECYT grant 1110569), MECESUP UAB0802, and the Research Office of Universidad Andres Bello (DI-35-11/R) (to D.P.-S.). S.B. was supported by a fellowship from the Ministry of Higher Education in Saudi Arabia.
Footnotes
Published ahead of print 6 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00901-13.
REFERENCES
- 1.McClane BA. 2007. Clostridium perfringens, p 423–444 In Doyle MP, Beuchat LR. (ed), Food microbiology: fundamentals and frontiers, 3rd ed. ASM Press, Washington, DC [Google Scholar]
- 2.McDonnell JL. 1986. Toxins of Clostridium perfringens type A, B, C, D, and E, p 477–517 In Dorner F, Drews J. (ed), Pharmacology of bacterial toxins. Pergamon Press, Oxford, United Kingdom [Google Scholar]
- 3.McClane B, Uzal FA, Miyakawa MF, Lyerly D, Wilkins T. 2006. The enterotoxic clostridia, p 698–752 In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes. Springer, New York, NY [Google Scholar]
- 4.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raju D, Setlow P, Sarker MR. 2007. Antisense-RNA-mediated decreased synthesis of small, acid-soluble spore proteins leads to decreased resistance of Clostridium perfringens spores to moist heat and UV radiation. Appl. Environ. Microbiol. 73:2048–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raju D, Waters M, Setlow P, Sarker MR. 2006. Investigating the role of small, acid-soluble spore proteins (SASPs) in the resistance of Clostridium perfringens spores to heat. BMC Microbiol. 6:50. 10.1186/1471-2180-6-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Setlow P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550–556 [DOI] [PubMed] [Google Scholar]
- 8.Paredes-Sabja D, Torres JA, Setlow P, Sarker MR. 2008. Clostridium perfringens spore germination: characterization of germinants and their receptors. J. Bacteriol. 190:1190–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paidhungat M, Ragkousi K, Setlow P. 2001. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J. Bacteriol. 183:4886–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moir A, Corfe BM, Behravan J. 2002. Spore germination. Cell. Mol. Life Sci. 59:403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffiths K, Zhang J, Cowan A, Yu J, Setlow P. 2011. Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol. Microbiol. 81:1061–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Setlow P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101:514–525 [DOI] [PubMed] [Google Scholar]
- 13.Setlow P. 2001. Resistance of spores of Bacillus species to ultraviolet light. Environ. Mol. Mutagen. 38:97–104 [DOI] [PubMed] [Google Scholar]
- 14.Moir A, Smith DA. 1990. The genetics of bacterial spore germination. Annu. Rev. Microbiol. 44:531–553 [DOI] [PubMed] [Google Scholar]
- 15.Paidhungat M, Setlow P. 2000. Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182:2513–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paidhungat M, Setlow P. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J. Bacteriol. 183:3982–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hudson KD, Corfe BM, Kemp EH, Feavers IM, Coote PJ, Moir A. 2001. Localization of GerAA and GerAC germination proteins in the Bacillus subtilis spore. J. Bacteriol. 183:4317–4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korza G, Setlow P. 2013. Topology and accessibility of germination proteins in the Bacillus subtilis spore inner membrane. J. Bacteriol. 195:1484–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paredes-Sabja D, Setlow P, Sarker MR. 2009. Role of GerKB in germination and outgrowth of Clostridium perfringens spores. Appl. Environ. Microbiol. 75:3813–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung JK, Keyburn AL, Carter GP, Lanckriet AL, Van Immerseel F, Moore RJ, Rood JI. 2010. The VirSR two-component signal transduction system regulates NetB toxin production in Clostridium perfringens. Infect. Immun. 78:3064–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Czeczulin JR, Collie RE, McClane BA. 1996. Regulated expression of Clostridium perfringens enterotoxin in naturally cpe-negative type A, B, and C isolates of C. perfringens. Infect. Immun. 64:3301–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannam TL, Rood JI. 1993. Clostridium perfringens-Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid 29:233–235 [DOI] [PubMed] [Google Scholar]
- 23.Kokai-Kun JF, Songer JG, Czeczulin JR, Chen F, McClane BA. 1994. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J. Clin. Microbiol. 32:2533–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duncan CL, Strong DH. 1968. Improved medium for sporulation of Clostridium perfringens. Appl. Microbiol. 16:82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paredes-Sabja D, Udompijitkul P, Sarker MR. 2009. Inorganic phosphate and sodium ions are cogerminants for spores of Clostridium perfringens type A food poisoning-related isolates. Appl. Environ. Microbiol. 75:6299–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabrera-Martinez RM, Tovar-Rojo F, Vepachedu VR, Setlow P. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J. Bacteriol. 185:2457–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Setlow B, Cowan AE, Setlow P. 2003. Germination of spores of Bacillus subtilis with dodecylamine. J. Appl. Microbiol. 95:637–648 [DOI] [PubMed] [Google Scholar]
- 28.Vepachedu VR, Setlow P. 2005. Localization of SpoVAD to the inner membrane of spores of Bacillus subtilis. J. Bacteriol. 187:5677–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart K-A, Yi X, Ghosh S, Setlow P. 2012. Germination protein levels and rates of germination of spores of Bacillus subtilis with overexpressed or deleted genes encoding germination proteins. J. Bacteriol. 194:3156–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelczar PL, Setlow P. 2008. Localization of the germination protein GerD to the inner membrane in Bacillus subtilis spores. J. Bacteriol. 190:5635–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bagyan I, Noback M, Bron S, Paidhungat M, Setlow P. 1998. Characterization of yhcN, a new forespore-specific gene of Bacillus subtilis. Gene 212:179–188 [DOI] [PubMed] [Google Scholar]
- 32.Stewart K-A, Setlow P. 2013. Numbers of individual nutrient germinant receptors and other germination proteins in spores of Bacillus subtilis. J. Bacteriol. 195:3575–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chirakkal H, O'Rourke M, Atrih A, Foster SJ, Moir A. 2002. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology 148:2383–2392 [DOI] [PubMed] [Google Scholar]
- 34.Paredes-Sabja D, Setlow P, Sarker M. 2011. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 19:85–94 [DOI] [PubMed] [Google Scholar]
- 35.Paredes-Sabja D, Setlow B, Setlow P, Sarker MR. 2008. Characterization of Clostridium perfringens spores that lack SpoVA proteins and dipicolinic acid. J. Bacteriol. 190:4648–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li YQ, Setlow B, Setlow P, Hao B. 2010. Crystal structure of the GerBC component of a Bacillus subtilis spore germinant receptor. Mol. Biol. 402:8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang G, Yi X, Li Y, Setlow P. 2011. Germination of individual Bacillus subtilis spores with alterations in the GerD and SpoVA proteins, which are important in spore germination. J. Bacteriol. 193:2301–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang G, Zhang P, Paredes-Sabja D, Green C, Setlow P, Sarker M, Li Y-Q. 2011. Analysis of the germination of individual Clostridium perfringens spores and its heterogeneity. J. Appl. Microbiol. 111:1212–1223 [DOI] [PubMed] [Google Scholar]
- 39.Kong L, Zhang P, Wang G, Yu J, Setlow P, Li Y-Q. 2011. Phase contrast microscopy, fluorescence microscopy, Raman spectroscopy and optical tweezers to characterize the germination of individual bacterial spores. Nat. Protoc. 6:625–639 [DOI] [PubMed] [Google Scholar]
- 40.Chen D, Huang SS, Li YQ. 2006. Real-time detection of kinetic germination and heterogeneity of single Bacillus spores by laser tweezers Raman spectroscopy. Anal. Chem. 78:6936–6941 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.