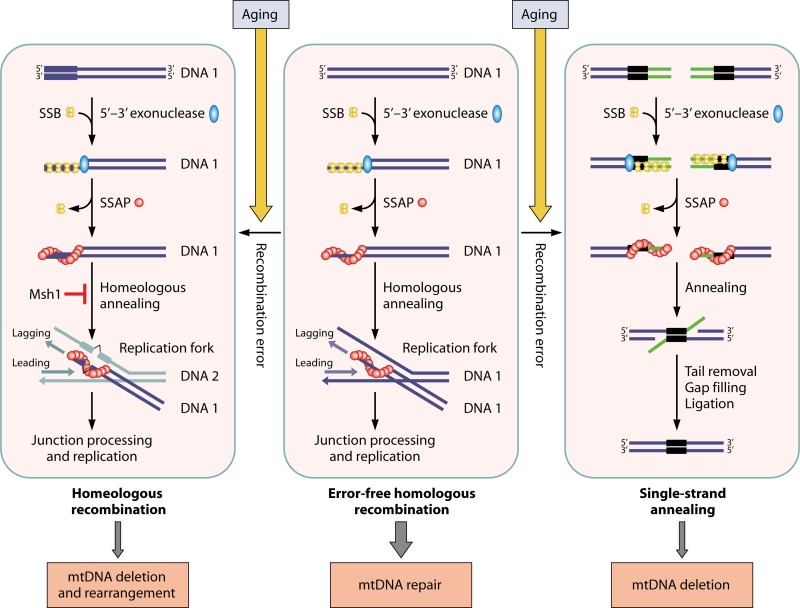
Fig 7.
Models for the operation mode of a recombination system of bacteriophage origin in mitochondria and its implications for the generation of mtDNA deletions in aged cells. A single-stranded DNA-annealing protein such as Mgm101 plays a central role in the repair of double-strand DNA breaks (middle panel). After the resection of a dsDNA end by a 5′-3′ exonuclease, SSB is bound to the exposed ssDNA to prevent the formation of secondary structures. SSB is then displaced by an SSAP like Mgm101. The SSAP/ssDNA nucleoprotein filaments initiate recombination by annealing to homologous single-stranded DNA on the lagging strand of a replication fork, like the Redβ protein in the phage λ (172, 173). The Mgt1/Cce1 endonuclease may process the recombination junction before the loading of the mtDNA replisome. However, recombinational errors may occasionally occur, either by a classic single-strand annealing between intramolecularly repeated sequences, which generates unrepairable mtDNA deletions (right panel), or by homeologous annealing to a nonhomologous lagging strand, which causes mtDNA deletions and rearrangements (left panel). The mismatch repair protein Msh1 may play a role in recombination editing by rejecting homeologous annealing. In young cells, mtDNA recombination is kept at a very low level to clear the rarely arising DSBs. In aged cells, mtDNA recombination increases as a result of oxidative stress and elevated mtDNA damage. This may inevitably increase erroneous recombinational events, which leads to the time-dependent accumulation of unfixable deleted and rearranged mtDNAs.