Abstract
Neisseria meningitidis is a commensal of humans that can colonize the nasopharyngeal epithelium for weeks to months and occasionally invades to cause life-threatening septicemia and meningitis. Comparatively little is known about meningococcal gene expression during colonization beyond those first few hours. In this study, the transcriptome of adherent serogroup B N. meningitidis strain MC58 was determined at intervals during prolonged cocultivation with confluent monolayers of the human respiratory epithelial cell line 16HBE14. At different time points up to 21 days, 7 to 14% of the meningococcal genome was found to be differentially regulated. The transcriptome of adherent meningococci obtained after 4 h of coculture was markedly different from that obtained after prolonged cocultivation (24 h, 96 h, and 21 days). Genes persistently upregulated during prolonged cocultivation included three genes (hfq, misR/phoP, and lrp) encoding global regulatory proteins. Many genes encoding known adhesins involved in epithelial adherence were upregulated, including those of a novel locus (spanning NMB0342 to NMB0348 [NMB0342-NMB0348]) encoding epithelial cell-adhesive function. Sixteen genes (including porA, porB, rmpM, and fbpA) encoding proteins previously identified by their immunoreactivity to sera from individuals colonized long term with serogroup B meningococci were also upregulated during prolonged cocultivation, indicating that our system models growth conditions in vivo during the commensal state. Surface-expressed proteins downregulated in the nasopharynx (and thus less subject to selection pressure) but upregulated in the bloodstream (and thus vulnerable to antibody-mediated bactericidal activity) should be interesting candidate vaccine antigens, and in this study, three new proteins fulfilling these criteria have been identified: NMB0497, NMB0866, and NMB1882.
INTRODUCTION
Neisseria meningitidis is a common colonist of the human nasopharyngeal epithelium, persisting in this location as a commensal organism for weeks or months (1, 2). From here, in a process that is poorly understood (but which apparently reflects human genetic predisposition, environmental factors, and the particular meningococcal strain present), it may rarely invade deeper tissues and the bloodstream to cause septicemia and meningitis (3–6). The dichotomous phenotypes of colonization and invasion are widely considered to be established early following the arrival of meningococci in the nasopharynx, with most invasive infections being thought to occur soon after this point. Late invasion is apparently rare (7). The interaction of meningococci with the nasopharyngeal epithelium has been a major driving force shaping the bacterial gene pool (8). While attention has understandably been focused largely on organisms isolated from the bloodstream, transcriptional changes promoting this phenotype cannot influence the gene pool, as these organisms are doomed, either through host bactericidal activity terminating infection or (all too often) through fatal infection removing organisms (and the host) from the pool. It is the bacterial transcriptome in organisms on the epithelial surface that not only determines the dynamics of the colonization process but also must be pivotal in the process of invasion. Transcriptomal studies focused on this phase of infection reveal changes during colonization that may increase or reduce the likelihood of invasion and identify critical bacterial determinants with vaccine potential. Published work in this area has been confined to studies which can identify only early events in the colonization process, having monitored bacterial/epithelial cell cocultivation for no longer than 6 h (9, 10). Here we report cocultivation of meningococci with human respiratory tract epithelial cells extended to a period of 21 days. The meningococcal transcriptome over this period was found to diverge markedly from the pattern found at 4 h, providing new insights into gene expression of potential relevance to invasive disease.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
N. meningitidis MC58 (serogroup B:15:P1.7.16b; ST-74) and its isogenic mutant strains (see below) were used. Meningococcal strains were routinely propagated at 37°C with 5% CO2 on GC agar (Difco) supplemented with 1% Vitox (Oxoid) (sGC) or at 37°C in brain heart infusion (BHI) broth (BD Biosciences) supplemented with 1% Vitox (sBHI broth).
Culturing of human epithelial cells.
The 16HBE14 bronchial epithelial cell line is derived from primary human bronchial epithelial cells transformed by the simian virus 40 (SV40) large T antigen (11). COR-L23 (catalogue no. 92031929) cells are derived from human lung carcinoma cells with epithelial morphology and were obtained from the European Collection of Cell Culture (Health Protection Agency, Porton Down, United Kingdom). The Chang epithelial cell line (ATCC CCL20.2) was obtained (in 2001) from the American Type Culture Collection (ATCC). The lung carcinoma cell line A549 (ATCC CCL-185) was obtained from the ATCC. The human tonsillar epithelial (HTE) cell line (transformed using human papillomavirus E6/E7) was a kind gift from Michael Apicella of the University of Iowa. All cell lines were propagated in Dulbecco's modified Eagle medium (DMEM) supplemented with 2 mM l-glutamine and 10% heat-inactivated fetal bovine serum (HIFBS) (all reagents from Invitrogen) with a humidified incubator at 37°C with 5% CO2.
Cocultivation model and control samples.
Epithelial cells grown to >90% confluence in either wells of 24-well plates (Cellstar; Greiner) or T75 flasks (BD Falcon; VWR) with permanently hydrophilic surfaces were used for determining meningococcal viable counts or preparing total RNA from adherent meningococci. For scanning electron microscopy (as well as for light microscopy using Giemsa staining), epithelial cells were grown on glass coverslips (13-mm diameter; British Drug House [BDH]) in 24-well plates. N. meningitidis cells grown on sGC plates were suspended in DMEM for initial inoculation at a multiplicity of infection of 30. The infected or noninfected (control) epithelial monolayers were incubated as specified above. Intermediate washes with phosphate-buffered saline (PBS) and the addition of fresh culture medium were carried out at 16-h intervals followed by 8-h intervals. The morphological integrity of epithelial monolayers was routinely monitored with an inverted microscope (Olympus). Representative monolayers were stained with trypan blue (Invitrogen) and viewed under the microscope. Images were photographed by using a C-35AD-4 (Olympus) camera (see Fig. S1 in the supplemental material).
RNA isolated from N. meningitidis cells cultured in DMEM for 3 h (DMEM3h) was used as a control sample. Bacteria grown on sGC plates were resuspended in supplemented DMEM to an optical density at 600 nm (OD600) of 0.2 in tissue culture flasks (T25) and incubated identically to the epithelial cell cocultivation preparations for 3 h.
Epithelial cell adherence and gentamicin protection assays.
At the desired cocultivation time points, infected monolayers were washed with PBS (three times with 1 ml per well), followed by incubation with 1% saponin (Sigma) for 10 min at 37°C (for the epithelial cell adherence assay). Bacteria as well as debris of human cells were separated by pipetting and plated onto sGC agar plates for CFU determinations. For the gentamicin protection assay, gentamicin (Sigma) was added to each sample at 150 μg/ml with supplemented DMEM, and the sample was incubated for 1 h at 37°C under 5% CO2. The resulting samples were further washed with PBS (3× 1 ml), and intracellular bacteria were released by using saponin and plated for enumeration.
Scanning electron microscopy.
At desired cocultivation time points, infected monolayers were washed as described above and incubated overnight at 4°C in 3% glutaraldehyde (Sigma), followed by a 30-min incubation with 1% osmium tetroxide (OsO4) (Sigma). Ethanol-dehydrated samples were processed by using a critical-point drying apparatus (Emitech K850) according to the manufacturer's instructions and subsequently mounted onto scanning electron microscopy stubs (JEOL) for further coating with palladium-gold using a Mini Sputter Coater (Polaron SC7620). The resulting samples were photographed by using a JEOL JSM-6390 scanning electron microscope.
RNA extraction.
Infected monolayers at 4 h, 24 h, 96 h, and 21 days in T75 flasks were washed with PBS and incubated with 0.5% trypsin containing 0.2% EDTA (Sigma) for 5 min at 37°C, followed by the addition of ice-cold PBS and pipetting (to separate cells and bacteria). The resulting mixture was centrifuged at 300 × g for 3 min in a prechilled centrifuge (Jouan) to pellet epithelial cells. The resulting supernatant or DMEM3h control samples were mixed with RNAprotect (Qiagen), incubated for 5 min at ambient temperature, and centrifuged for 10 min at 3,000 × g. The pellets were subjected to RNA extraction by using the FastPrep FP120 system (MP Biochemicals) according to the manufacturer's instructions. The RNA samples were further treated with DNase (Invitrogen), purified by using an RNeasy kit (Qiagen), and analyzed by using PCR and a Bioanalyzer (Agilent) to monitor RNA quality and contamination of human RNA and bacterial genomic DNA (gDNA). RNA samples selected from each biological group were tested for contamination of genomic DNA by using primers for the meningococcal gene NMB0152 (see Table S1 in the supplemental material), the HotStar PCR kit (Qiagen), and a DNA Engine thermal cycler (Bio-Rad) for 40 cycles, according to the manufacturer's recommendations. One-fifth of each PCR mixture was examined by agarose gel electrophoresis and ethidium bromide staining and visualized on a UV transilluminator. No PCR product of the expected molecular weight (that of positive-control MC58 genomic DNA as the template) was detected (data not shown). There was no human RNA or genomic DNA contamination, as measured by Bioanalyzer analysis, i.e., no distinct peaks observed at a high molecular weight (see Fig. S2 in the supplemental material).
Microarray hybridization and analysis.
Labeling of RNA (1 μg) and genomic DNA (0.1 μg) obtained from MC58 was performed as previously described (12). Briefly, RNA samples were labeled by using random primers, a Superscript III kit (Invitrogen), and Cy5 (GE Healthcare), with an initial sample denaturation step at 72°C and a subsequent elongation step at 42°C for >2 h. DNA samples were labeled by using random primers, a large-fragment DNA polymerase I kit (Invitrogen), and Cy3 (GE Healthcare), with an initial sample denaturation step at 95°C and a subsequent elongation step at 37°C. The pan-Neisseria meningitidis array NMGv2.2.0 was designed by the Bacterial Microarray Group at St. George's University of London (BμG@S) (http://bugs.sgul.ac.uk/) and manufactured on the Agilent SurePrint 8×15 K platform (Agilent), with each gene being represented by three different oligonucleotides (40- to 60-mers). Each array slide was cohybridized with one of the Cy5-labeled RNA samples and Cy3-labeled gDNA (universal reference) by using the Agilent hybridization kit and microarray hybridization oven with rotating hybridization chambers. The resulting slides were washed by using an Oligo aCGH & ChIP-on-Chip wash buffer kit (Agilent) and scanned by using a high-resolution microarray scanner (Agilent). Data were extracted and processed from each scanned image by using Agilent Feature Extraction v10.7.3.1 (Agilent) and further analyzed by using GeneSpring software (Agilent).
Normalization of microarray data and comparison of transcriptomes between time points, e.g., 4 h versus 3 h with DMEM treatment, were performed as previously described (12). Briefly, the signal intensities resulting from hybridization of Cy5-labeled cDNA derived from meningococcal RNA were normalized to the signal intensities from Cy3-labeled genomic DNA on each spot (per-spot normalization). The normalized spot values of technical replicates for each gene were averaged and normalized to the 50th percentile (median) values of each array with locally weighted scatterplot smoothing (LOWESS) applied for smoothing using 20% of the data (per-gene/per-array normalization). To identify genes differentially expressed between DMEM3h (control) and other biological groups (4 h, 24 h, 96 h, and 21 days), the normalized intensities for each gene derived from averaged biological replicates within a group were compared by using analysis of variance (ANOVA). Cutoff values of a 1.5-fold change (P < 0.01, corrected for multiple testing using Benjamini-Hochberg correction) were used. Correlation of gene expression profiles was calculated as previously described (13). Principal-component analysis (PCA) was performed by using all genes that passed quality control testing and default settings in GeneSpring. Gene clustering was done with GeneSpring using a hierarchical algorithm, and transcriptome similarity was measured by Euclidean distance.
DAVID (14) was used to identify differentially regulated gene groups according to their associated functional annotations. Benjamini multiple correction (P < 0.01) was used to select the most significantly enriched functional groups.
Quantitative real-time reverse transcription-PCR (RT-PCR).
RNA samples from three fresh biological replicates were prepared as described above and were reverse transcribed to first-strand cDNA by using Superscript III and random primers (Invitrogen). Real-time PCR was performed as described previously (12), using TaqMan technologies, Universal PCR Master Mix, and the StepOnePlus system (Applied Biosystems). The primers were designed and manufactured by using Assay-by-Design (Applied Biosystems) and are listed in Table S1 in the supplemental material. The reporter dye and quencher were 6-carboxyfluorescein (FAM) and NFQ, respectively. We used 10 ng total RNA per reaction. The reference gene was 16S rRNA. Statistical analysis was based on methods reported previously by Livak and Schmittgen (15), and detailed statistical formulations are described in Applied Biosystems user bulletin no. 2 (P/N4303859).
Construction and characterization of meningococcal mutants.
Genes in the locus spanning NMB0342 to NMB0348 (NMB0342–NMB0348 locus) share identical sequences with homologous genes in N. meningitidis strain 8013 (16, 17). Strain MC58 was transformed with genomic DNA from the corresponding isogenic mutants of strain 8013 (kindly provided by Vladimir Pelicic of the Imperial College London) made through transpositional mutagenesis (18). A backcross transformation strategy previously used in meningococcal studies (19) was used to ensure that the phenotypic changes in each mutant strain were due to the mutation of the targeted gene rather than the incorporation of unintended mutations at other chromosomal loci. It must be conceded that, although very unlikely, it is not impossible that despite this backcross approach, undetectable recombination may still have occurred in other loci. Briefly, DNA from first-generation (kanamycin-resistant) mutants was used to retransform the kanamycin-sensitive parental strain (MC58) to generate second-generation mutants. The resulting transformants, grown on sGC plates containing kanamycin (50 μg/ml), were selected for PCR amplification using gene-specific primers (see Table S1 in the supplemental material) to confirm insertion of a Kanr cassette in the targeted genes NMB0342, NMB0344, NMB0345, NMB0347, and NMB0348 (see Fig. S3 in the supplemental material). The PCR products were sequenced by using the BigDye reaction kit (Applied Biosystems) and an ABI3730XL sequencer (Applied Biosystems).
For bacterial growth curves, N. meningitidis cells (MC58 and its isogenic mutants) were grown for 16 h on sGC plates, harvested, and diluted in sBHI broth or DMEM supplemented with 10% HIFBS to give an initial OD600 of 0.2. The resulting bacterial suspension was incubated at 37°C with gentle shaking, and OD600 readings were taken intermittently.
Expression of targeted or adjacent genes was studied by using RT-PCR. RT was performed by using RNA extracted from each mutant grown on sGC plates, the SuperScript III kit (Invitrogen), and reverse primers (see Table S1 in the supplemental material). The resulting cDNA was used as the template for PCR using the HotStar PCR kit (Qiagen). The primers used for RT-PCR are listed in Table S1 in the supplemental material.
Statistical significance was calculated for the differences between adherent CFU obtained from wild-type strain MC58 and those obtained from each of its isogenic mutant strains by using the two-tailed Student t test.
Nucleotide sequence accession numbers.
The array design is available at BμG@Sbase (accession no. A-BUGS-45 [http://bugs.sgul.ac.uk/]) and also at ArrayExpress (accession no. A-BUGS-45 [http://www.ebi.ac.uk/microarray-as/ae/]). Fully annotated microarray data have been deposited in the BμG@Sbase database (accession no. E-BUGS-141) and also in the ArrayExpress database (accession no. E-BUGS-141), which is in compliance with minimum information about a microarray experiment (MIAME) protocols.
RESULTS
Establishing and characterizing a meningococcal epithelial colonization model.
We sought to establish a meningococcal/human epithelial cell cocultivation model using immortalized epithelial cells that would support meningococcal serogroup B strain MC58 adherence for a long period and at a sufficient bacterial density to yield enough meningococcal RNA (ca. 1 μg/sample) for transcriptome analysis. Five cell lines were tested in a system of confluent culture in flat-bottomed flasks, with twice-daily washing and replenishment of culture medium (DMEM) (see Fig. S4 in the supplemental material). At 28 h, there were significant numbers of meningococci adherent to 16HBE14 and COR-L23 cells but no detectable meningococci on Chang, A549, or transformed human tonsillar epithelial (HTE) cells (data not shown for the latter two lines). 16HBE14, the SV40-immortalized bronchial epithelial cell line used by Grifantini et al. (10) for their transcriptomal study, was the only cell line supporting meningococcal adherence at 48 h cocultivation and beyond, with the CFU per monolayer being comparable at 28, 48, and 96 h but with a further increase at 21 days (see Fig. S4 in the supplemental material). During cocultivation, epithelial cells were examined daily for gross signs of cell detachment from the plastic surface, and cell viability was examined periodically by trypan blue staining (see Fig. S1 in the supplemental material). Giemsa staining revealed that by 48 h, the majority of COR-L23 cells had lost their nucleoli, with no meningococci to be seen on the cell surface (Fig. 1A). In contrast, 16HBE14 cells retained distinct nucleoli within nuclei, and numerous adherent meningococci were present (Fig. 1B). The 16HBE14 cell line was the only cell line that could survive an extended period of cocultivation with N. meningitidis strain MC58 under our experimental conditions and therefore was selected for further study.
Fig 1.
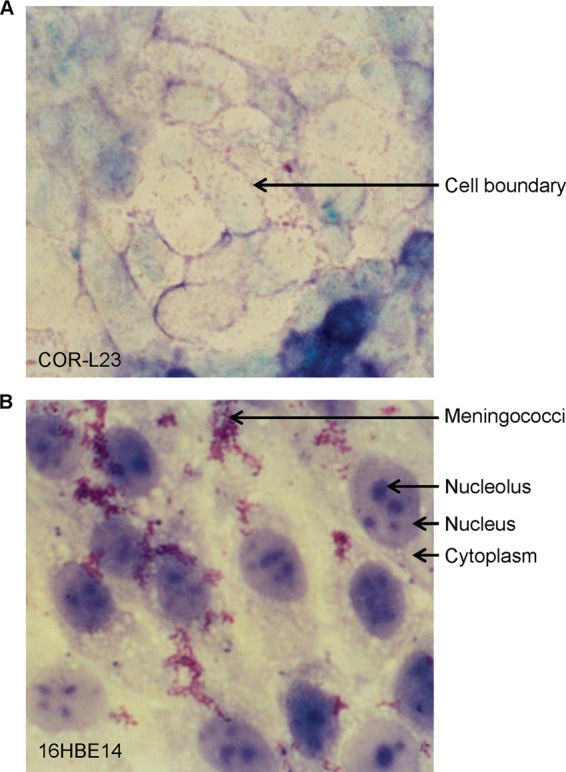
Viability of epithelial cells cocultivated with N. meningitidis MC58. Epithelial cell monolayers cocultivated with bacteria for 48 h were washed and stained with Giemsa. (A) COR-L23 cells at a ×400 magnification. The arrow indicates an example of a cell boundary. (B) 16HBE14 cells at a ×1,000 magnification. Arrows indicate examples of cytoplasm, nucleolus, nucleus, and a group of meningococci.
Meningococcus-16HBE14 cell interactions were characterized during extended periods of cocultivation by scanning electron microscopy. After 10 h, meningococci were present as microcolonies, and the surface of the epithelial cells was marked by short microvillus-like membrane protrusions (20, 21) (Fig. 2A). After 48 h, elongated filopodium-like protrusions were also observed (20, 21), entangled with the meningococcal microcolonies (Fig. 2B), as previously described (22). After 21 days, meningococci were found to be extensively entangled with filopodium-like membrane protrusions, with increased numbers of straight, fine, pilus-like filaments connecting bacteria (Fig. 2C). Comparison with images obtained at earlier time points (10 h and 48 h) indicates that the number of meningococci at 21 days was increased, which correlates with the increased number of adherent CFU at 21 days of cocultivation (see Fig. S4 in the supplemental material).
Fig 2.
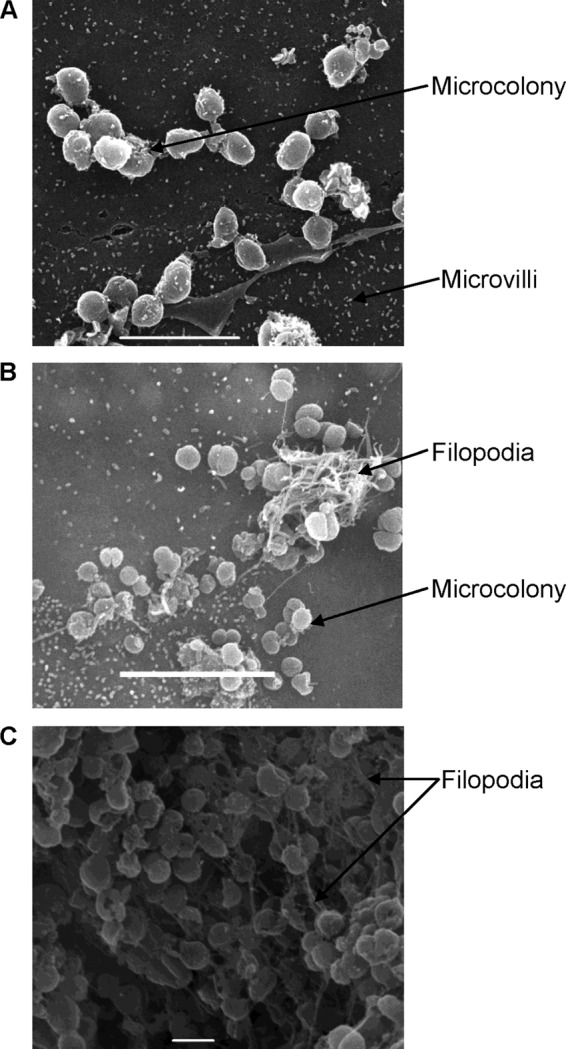
Scanning electron microscopy images of 16HBE14 cells cocultivated with N. meningitidis MC58. Images were taken at 10 h (A), 48 h (B), and 21 days (C) of cocultivation, with white bars indicating 5, 5, and 1 μm, respectively.
Reliability and comparability of transcriptomes.
RNA samples prepared from adherent meningococci at each of four cocultivation time points (4 h, 24 h, 96 h, and 21 days) were reverse transcribed to cDNA, labeled, and hybridized to pan-N. meningitidis DNA arrays. Four biological replicates were made at each time point except the last, at which time there were 3 replicates. RNA samples from meningococci grown to mid-exponential phase (3 h in DMEM) were processed similarly and hybridized to the meningococcal DNA array to generate a reference transcriptome for later comparisons (termed the DMEM3h transcriptome). Transcriptomes at each time point were compared. Replicates for each time point were closely correlated, ranging from 83% to 99% (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). Correlations between time points ranged between 62% and 99% (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). The correlation between DMEM3h (control) and other samples ranged between 68 and 85%. The highest correlations were found, as expected, between biological replicates; the next highest correlation was found between transcriptomes at 24 h and 96 h, which was in turn higher than those between these time points and 4 h, with the 21-day transcriptomes being the least similar to those at earlier time points. This is interpreted as there being substantial transcriptional changes occurring early, followed by a period of relative transcriptional consistency before late changes supervene, plausibly reflecting a biologically relevant model of epithelial colonization. Transcriptomes from individual biological replicates were also compared by using principal-component analysis (PCA). PCA grouped biological replicates into three main clusters, and within each cluster, transciptomes for each time point group were closely related (see Fig. S5 in the supplemental material), which was in agreement with the correlation analysis (see the data at http://bugs.sgul.ac.uk/E-BUGS-141).
Overall transcriptome comparison between cocultivation time points: the transcriptome at 4 h was markedly different from those at prolonged coincubation times.
Meningococcal transcriptomes recovered at each time point were consolidated and analyzed by using PCA. Transcriptomes at all meningococcal/epithelial cocultivation time points were clearly separated from the DMEM3h transcriptome with respect to both PCA components 1 and 2, representing 37% and 28.5% variances, respectively (Fig. 3A). Transcriptomes determined at the four cocultivation time points were shifted mainly along PCA component 2, with those at 4 h and 21 days being positioned at the top and bottom of the scale and those at 24 h and 96 h being clustered together at an intermediate position (Fig. 3A).
Fig 3.
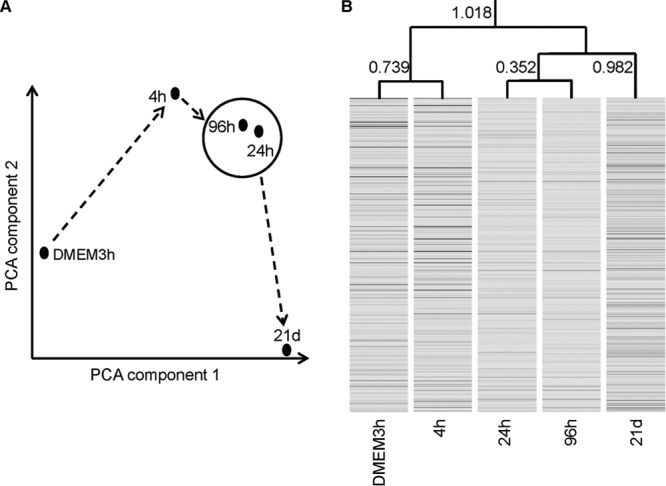
Comparison of meningococcal transcriptomes obtained at different cocultivation time points. (A) PCA. Solid dots in the plot represent the relative positions of samples at different cocultivation time points. Dashed arrows represent the passage of time. (B) Gene clustering with similarity between time points measured by Euclidean distance, as indicated.
The four consolidated transcriptomes derived from cocultures were compared individually with the DMEM3h transcriptome (4 pairwise comparisons, e.g., 4 h versus DMEM3h), seeking significant differences in individual gene expression. A total of 819 genes that showed a >1.5-fold change with P values of <0.01 (after correcting for multiple comparisons) for at least 1 of the 4 pairwise comparisons were identified as being differentially expressed at various time points (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). (From here on, the terms up- and downregulated refer to these differences.) Using this selected set of genes and a clustering algorithm, we further established the Euclidean distance relation between transcriptomes recovered at different time points (Fig. 3B). Transcriptomes at 24 h and 96 h were most similar, with a distance of 0.352, while that at 4 h was closest to the reference at 3 h in DMEM (distance = 0.739). The transcriptome at 21 days was not similar to that at any other time point but was closer to the combined group at 24 h and 96 h (distance = 0.928) than anything else.
A total of 160, 262, 267, and 264 genes were upregulated, and 222, 210, 182, and 288 genes were downregulated, at 4 h, 24 h, 96 h, and 21 days, respectively; between 7% and 14% of 2,062 genes on our array platform were differentially regulated at any time point (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). A total of 230 genes were upregulated, and 164 genes were downregulated, at both 24 h and 96 h (Fig. 4). A total of 49 and 54 genes were up- and downregulated, respectively, at all time points from 4 h to 21 days compared to transcription after 3 h of incubation in DMEM (see the data at http://bugs.sgul.ac.uk/E-BUGS-141).
Fig 4.
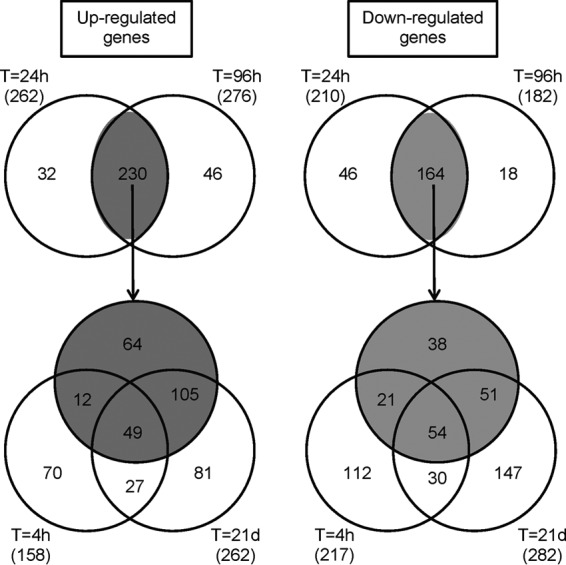
Venn diagrams showing numbers of differentially regulated genes coidentified at different cocultivation time points. The shaded circles in three-way Venn diagrams represent 230 up- and 164 downregulated genes coidentified at 24 h and 96 h (in two-way Venn diagrams). The total numbers of differentially regulated genes for each time point are indicated in parentheses.
Differentially expressed genes were grouped functionally into Clusters of Orthologous Groups (COGs), defined by the U.S. National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/) (see Fig. S6 in the supplemental material). Genes belonging to the groups “transcription,” “translation,” “posttranslational modification,” and “cell wall/membrane biogenesis,” mostly downregulated at 4 h, had different patterns of transcription at later times (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). There were 105 and 51 genes up- and downregulated, respectively, during prolonged (24 h, 96 h, and 21 days) coincubation (Fig. 4). Among the differentially expressed genes, three were identified as transcriptional regulators, hfq (NMB0748), phoP (also termed misR) (NMB0595), and lrp (NMB1650), all upregulated at later time points (24 h, 96 h, and 21 days), compared to a lower level of expression at 4 h (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). This suggests that there is substantial, genome-wide resetting of transcriptional activity in meningococci during a prolonged interaction with epithelial cells.
Up- and downregulated genes at different time points were further analyzed by using DAVID (14), a program for gene enrichment analysis according to associated functional annotations. Significant gene enrichment (P < 0.01 with Benjamini correction) for 5 functional groups was found in all upregulated gene lists (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). All 5 functional groups were associated with bacterial surface structures at all time points. A further two were enriched in surface structures only at 21 days (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). Among downregulated genes at 21 days, 11 functional groups were identified as being significantly enriched, with most being related to DNA modification (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). Here we focus on those meningococcal genes which encode proteins involved directly in epithelial interaction or encode outer membrane proteins with vaccine potential.
Genes involved in epithelial adhesion and invasion.
Two groups of adhesins, type IV pili and opacity proteins, are considered to be crucial for adherence of meningococci to human epithelial cells (23). The pilE (NMB0018) gene, which encodes the major pilin subunit, together with 7 pilS genes (NMB0019, NMB0020, and NMB0022 to NMB0026), was upregulated at all time points (Fig. 5) (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). Upregulation of pilE was marginal (1.7-fold) at 4 h but much higher at later time points (6.6- to 12-fold) (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). Other genes involved in pilus biogenesis (24, 25) were mostly not differentially regulated. Of the 25 genes encoding products potentially involved in pilus biogenesis and function, pilP (NMB1811), pilV (NMB0547), and pilN (NMB1809) were downregulated, and pilT (NMB0768) was upregulated, at various time points (see the data at http://bugs.sgul.ac.uk/E-BUGS-141).
Fig 5.
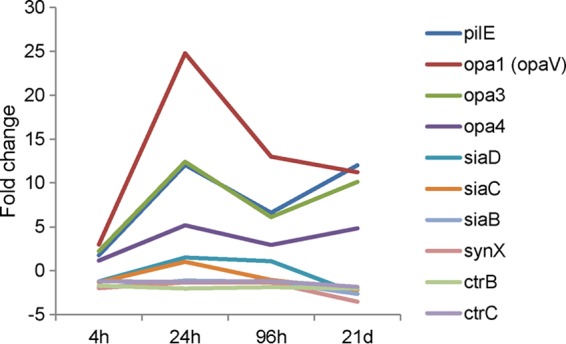
Transcriptional profiles of selected genes. Levels of meningococcal gene expression at different cocultivation time points (as indicated on the x axis) and for DMEM at 3 h were compared and are expressed as fold changes.
opa1 (opaV) (NMB0042), opa3 (NMB1465), and opa4 (NMB1636), encoding opacity proteins previously shown to be involved not only in adhesion but also in invasion of epithelial cells (26), were upregulated at all time points (Fig. 5) (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). Genes encoding other known adhesins (23), opcA (NMB1053), nhhA (NMB0992), app (NMB1985), nadA (NMB1994), hrpA1 (NMB0497), and hrpA2 (NMB1779), were not differentially expressed. mspA (NMB1998) was downregulated at all time points (see the data at http://bugs.sgul.ac.uk/E-BUGS-141).
NMB1965, encoding the l-glutamate transporter GltT, involved in epithelial cell invasion and intracellular survival (27, 28), was upregulated at 96 h (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). NMB1963, encoding a putative periplasmic transporter functioning on the same pathway and likely to be involved in epithelial cell adhesion/invasion (27), was upregulated at 24 h and 96 h (see the data at http://bugs.sgul.ac.uk/E-BUGS-141).
NMB0964, encoding the TonB-dependent zinc receptor ZnuD, involved in epithelial adhesion and invasion (29), was upregulated at 4 h and 96 h (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). NMB1882, also encoding a TonB-dependent receptor, was upregulated at 96 h but downregulated at 21 days (see the data at http://bugs.sgul.ac.uk/E-BUGS-141).
The majority of genes involved in meningococcal capsular polysaccharide biosynthesis and transport (NMB0067 to NMB0070, NMB0072, and NMB0073), believed to negatively influence adherence and invasion of human epithelial cells (30), were downregulated at various time points (Fig. 5) (see the data at http://bugs.sgul.ac.uk/E-BUGS-141).
Expression of different lipooligosaccharide (LOS) phenotypes influences not only meningococcal survival in human blood (31) but also interactions with epithelial surfaces (32, 33). Genes involved in LOS biosynthesis, lgtE (NMB1926), lgtA (NMB1927), lgt (NMB1072), lpxK (NMB0672), and lpxA (NMB0178), were downregulated at various time points (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). Genes involved in LOS transport (34) were not differentially expressed, except for lptB (NMB0356), which was marginally upregulated at 24 h and 96 h (see the data at http://bugs.sgul.ac.uk/E-BUGS-141).
The products of NMB0342 and NMB0345 have been implicated in meningococcal epithelial cell adhesion, survival in human blood, and virulence (35, 36). NMB0345 is recognized by sera obtained from individuals colonized by meningococci (2). These genes and other adjacent genes form the cluster NMB0342–NMB0348, all upregulated during prolonged coincubation with 16HBE14 epithelial cells, especially at 24 h and 96 h (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). We have chosen to characterize this locus further.
The NMB0342–NMB0348 locus.
Mutants were constructed in strain MC58 through insertion of a kanamycin-resistant cassette into the open reading frame of NMB0342 (encoding intracellular septation protein A), NMB0344 (encoding a BolA/YrbA family protein), NMB0345 (encoding a predicted cell binding factor), NMB0347 [encoding d-tyrosyl-tRNA(Tyr) deacylase], and NMB0348 (encoding tRNA-dihydrouridine synthase A). Semiquantitative RT-PCR was performed to investigate the effect of each mutation on the expression of downstream genes; no polar effects were demonstrated (see Fig. S7 in the supplemental material).
There was no difference in growth between any of the mutants and wild-type strain MC58 in the DMEM used for cocultivation of meningococci with epithelial cells (see Fig. S8 in the supplemental material). The resulting mutants, as well as wild-type strain MC58, were tested for their ability to adhere to 16HBE14 cell layers at 4 h, 24 h, and 96 h. In comparison with MC58, each mutant strain demonstrated significantly (P < 0.02) reduced adherence, with the differences increasing at 24 h and 96 h (Fig. 6).
Fig 6.
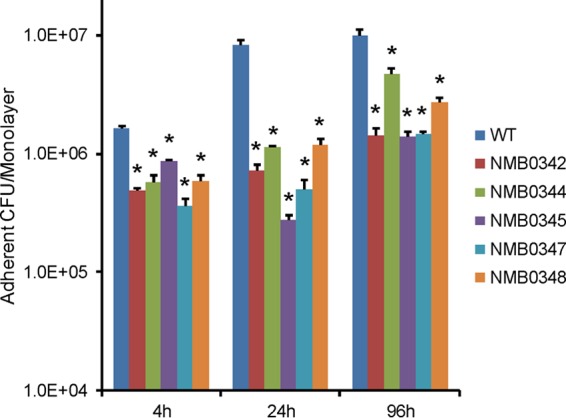
Epithelial cell adherence assay of meningococcal mutants. Monolayers of 16HBE14 cells were incubated with wild-type (WT) N. meningitidis MC58 and its isogenic mutants (indicated as NMB0342, NMB0344, NMB0345, NMB0347, and NMB0348, for the genes being disrupted) for 4 h, 24 h, and 96 h. Numbers of adherent meningococcal CFU were obtained from three biological replicates. Error bars indicate standard errors of the means. Asterisks indicate CFU with a significant difference (P < 0.02) between the corresponding mutant and the wild type.
Transcription of NMB0342, NMB0344, and NMB0345 was investigated by using quantitative real-time RT-PCR. Independent biological samples were prepared from meningococci (MC58) harvested after 24 h and 96 h of cocultivation with 16HBE14 epithelial cell layers and compared to samples from bacteria incubated for 3 h in DMEM alone. All three genes were upregulated, between 1.7- to 4.5-fold at both time points (Fig. 7).
Fig 7.
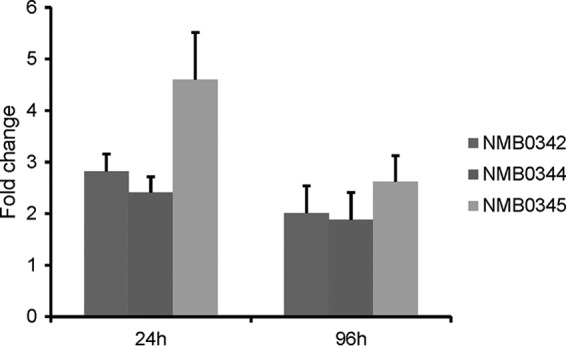
Comparison of gene expression using real-time RT-PCR. Independent biological RNA samples were prepared from MC58 cells harvested after 24 h and 96 h of cocultivation with 16HBE14 epithelial cells and compared to RNA samples from MC58 cells incubated for 3 h in DMEM alone (DMEM3h). The results of comparisons are expressed as fold changes. Error bars indicate standard errors of the means calculated from three biological replicates.
Genes encoding surface antigens with vaccine potential.
Sixteen genes encoding proteins identified by PSORTb (37) to be located in the bacterial outer membrane (and hence to have vaccine potential) were found to be differentially regulated (up- or downregulated) in the present study (Table 1). They included six genes which were previously investigated as meningococcal vaccine candidates. These genes are NMB1541 (encoding the lactoferrin binding protein LbpB) and NMB0460 (encoding the transferring binding protein TbpB) (38–40), which were downregulated at all time points, and genes encoding PorA (NMB1429), PorB (NMB2039), and RmpM (NMB0382), recently shown to be present as protein complexes (41), which shared a different expression pattern: downregulated at 4 h and upregulated at later time points. NMB1882, identified as a candidate vaccine antigen on the strength of being upregulated during incubation in nonbactericidal human blood (13), was upregulated upon coincubation with 16HBE14 cells by 96 h but downregulated at 21 days.
Table 1.
Differentially expressed genes encoding outer membrane proteinsa
| Gene identification | Gene name | FC at each cocultivation time point |
|||
|---|---|---|---|---|---|
| 4 h | 24 h | 96 h | 21 days | ||
| NMB0345 | 4.1 | 3.0 | |||
| NMB0382 | rmpM | −2.7 | 4.8 | 2.6 | 2.7 |
| NMB0460 | tbpB | −1.6 | −1.8 | −1.7 | −2.8 |
| NMB0497 | −2.1 | −2.6 | −2.8 | −3.1 | |
| NMB0866 | −2.7 | −2.5 | −1.9 | −2.3 | |
| NMB1405 | −2.4 | ||||
| NMB1429 | porA | −2.8 | 5.0 | 4.2 | 2.3 |
| NMB1541 | lbpB | −1.5 | −1.8 | −1.7 | −2.3 |
| NMB1567 | 3.1 | 2.8 | 1.7 | ||
| NMB1768 | 1.7 | 1.6 | 1.7 | ||
| NMB1882 | 1.5 | −1.7 | |||
| NMB1969 | ausP | −1.7 | −1.7 | −1.7 | −1.7 |
| NMB1988 | fetA | −1.6 | |||
| NMB1998 | mspA | −1.8 | −1.6 | −1.5 | −1.7 |
| NMB2039 | porB | −4.0 | 3.7 | 2.4 | 4.2 |
| NMB2050 | −2.0 | −2.2 | |||
Two further proteins of unknown function listed in Table 1, NMB0866 and NMB0497, were downregulated at all time points upon coincubation with 16HBE14 cells but were found previously by Hedman et al. (13) to be upregulated after 20 and 240 min of incubation in nonbactericidal human blood. Three more genes, NMB1405, NMB1988, and NMB2050, also downregulated at various time points in the present study, were also downregulated in blood (13).
NMB0345, NMB1567, and NMB1768, all encoding proteins of unknown function, were upregulated at various time points during prolonged cocultivation with 16HBE14 cells, while NMB1969 and NMB1998, encoding autotransporters (13, 42, 43), were downregulated at all time points.
Grifantini et al. (10) carried out a short time course study otherwise comparable to ours, coincubating MC58 with 16HBE14 cells for up to 3 h. They identified over 50 genes that were highly upregulated at at least one time point in this period and investigated 9 of these genes by fluorescence-activated cell sorter (FACS) analysis. Two of the nine genes, NMB0787 and NMB0995, were also upregulated in the present study, in each case not at 4 h but at 24 and 96 h and still at 21 days.
Williams et al. (2) identified 43 vaccine candidate genes through immunoreactivity of their products with sera obtained from individuals colonized by serogroup B meningococci. Sixteen of these genes were found to be upregulated in the present study (Table 2), with 15 of 16 being upregulated at all three later time points (24 h, 96 h, and 21 days), including porA, porB, rmpM, and groEL, previously found to also be highly immunoreactive to sera from immunized mice (41) or from convalescent meningococcal patients (44) in later studies. PorA is one component of the investigational serogroup B meningococcal vaccine 4CMenB (Bexsero) (45). However, genes encoding the three other major antigenic components, factor H binding protein (NMB1870), neisserial heparin binding antigen (NMB2132), and the adhesin NadA (NMB1994), were not found to be differentially expressed in our study.
Table 2.
Differentially expressed genes encoding immunoreactive proteinsa
| Gene identification | Gene name | FC at each cocultivation time point |
|||
|---|---|---|---|---|---|
| 4 h | 24 h | 96 h | 21 days | ||
| NMB0018 | pilE | 1.8 | 12.1 | 6.6 | 12.0 |
| NMB0128 | rplA | −2.2 | 4.0 | 3.6 | 1.7 |
| NMB0143 | rplD | −1.6 | 10.0 | 7.3 | 1.7 |
| NMB0345 | 4.1 | 3.0 | |||
| NMB0382 | rmpM | −2.7 | 4.8 | 2.6 | 2.7 |
| NMB0546 | adhA | −2.6 | 1.8 | ||
| NMB0554 | dnaK | 2.5 | |||
| NMB0604 | 2.1 | 2.2 | |||
| NMB0634 | fbpA | 3.5 | 2.5 | ||
| NMB1313 | tig | 2.1 | 1.6 | 1.6 | |
| NMB1429 | porA | −2.8 | 5.0 | 4.2 | 2.3 |
| NMB1533 | 1.8 | ||||
| NMB1710 | gdhA | 3.5 | 2.9 | 1.5 | |
| NMB1972 | groEL | 1.9 | 1.5 | 2.0 | |
| NMB2039 | porB | −4.0 | 3.7 | 2.4 | 4.2 |
| NMB2159 | gapA-2 | 2.0 | 2.8 | ||
Immunoreactive proteins were previously identified by using sera from individuals colonized with serogroup B meningococci (2). Only values with a P value of <0.01 are shown.
DISCUSSION
Published transcriptomic studies of meningococci adherent to epithelial cells have until now been confined to short periods of cocultivation (up to 6 h) (9, 10). However, it is well known that meningococci colonize the human nasopharynx for weeks to months (1, 2), and therefore, such short-term adherence studies cannot throw light on the importance of meningococcal gene products that act later in the process of host-pathogen interactive biology. Here we have investigated differences over time in the transcriptome of serogroup B meningococci adherent to cultured respiratory epithelial cells in a prolonged-colonization model. Principal-component analysis indicates that transcriptomes obtained at 24 h, 96 h, and 21 days are quite different from that seen at 4 h (Fig. 3A). Transcriptomes at 24 h and 96 h appeared to be very similar to each other but substantially different from that at 21 days (Fig. 3B), comparisons emphasized in Venn diagrams of differentially expressed genes at the three different time points (Fig. 4). Since it is generally considered that where invasive disease occurs, meningococci invade soon after they arrive in the nasopharynx, the transcriptome at relatively early cocultivation time points may be more representative of the biological state of meningococci poised for invasion than that seen after prolonged cocultivation. In contrast, sustained upregulation upon prolonged cocultivation may identify a meningococcal gene set relevant to the commensal state.
Comparing our results with those of previously reported studies of short-term cocultivation, 107 of the 347 meningococcal genes identified by Grifantini et al. (10) as being differentially regulated over 3 h of contact with 16HBE14 cell monolayers were also found in our differential gene lists (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). Of 55 genes that Grifantini et al. (10) found to be substantially upregulated at some point in the 180 min of their experiment, 18 were similarly expressed in our study. Seven were differentially expressed in the opposite direction, while the remaining 30 did not show significant differential expression at our time points (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). Differences in methodology, and in particular in the microarrays used, growth conditions for control samples, and the statistical algorithm employed, require caution in the interpretation of such comparisons. Agreement between studies should be seen more as hypothesis strengthening rather than proving a causative relationship between specific gene expression and biology. However, it is interesting to note that three genes involved in sulfur acquisition and metabolism (cysD, cysJ, and cysN), identified as being upregulated in the study by Grifantini et al. (10) and hypothesized to encode products involved in the epithelial infection process, were also upregulated in our study.
Grifantini et al. (10) used FACS analysis to detect meningococcal proteins on the bacterial surface, comparing adhering and nonadhering bacteria. NMB0787 and NMB0995, encoding surface-expressed proteins upregulated in adherent organisms, were also upregulated in our study.
In a second reported short-term cocultivation study, Dietrich et al. (9) found 67 meningococcal genes upregulated during a 6-h cocultivation with HEp-2 cell monolayers. Twenty-one of these genes were also found to be upregulated in our gene list (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). However, differences in experimental settings, especially with different cell lines being used, likely account for the disparities between the present study and that of Dietrich et al. (9).
The modest correlation of our results with the results of those two studies should be seen in the context of the fact that only 6 genes identified as being substantially upregulated by Grifantini et al. (10) were also identified in the list of differentially expressed genes in the study by Dietrich et al. (9): NMB0517, NMB0787, NMB0881, NMB0994, NMB0995, and NMB1017. It is noteworthy that two of these six genes (NMB0787 and NMB0995) in the set identified by Grifantini et al. (10) to be surface exposed in epithelial cell-adherent meningococci were highly upregulated in our study after cocultivation with 16HBE14 monolayers for >24 h (fold change [FC], 2.4 to 7.4) although not differentially expressed at 4 h. Focusing on NMB0995, Kuwae et al. (46), in agreement with our results, reported upregulation following infection of a human pharyngeal epithelial cell line only after 6 h, a protein not detected on the meningococcal surface at the early adhesion stage (2 h). The gene, termed nafA (for neisserial antiaggregation factor A) by those authors, encodes a surface-expressed protein which interacts with components of the meningococcal type IV pilus to prevent excess autoaggregation and the formation of microcolonies on epithelial surfaces during colonization. It is highly conserved in sequence between meningococcal strains, a nafA mutant is less virulent in mice following intraperitoneal challenge, and purified protein elicits bactericidal antibodies in mice (10, 46); these factors all establish its vaccine candidacy. Extending such studies, we have shown previously that nafA is upregulated in meningococci cocultivated with nonbactericidal fresh human blood (13).
In a third study, a serogroup B meningococcal carriage strain (alpha-710) or MC58 was cocultivated with FuDa epithelial cells for 6 h, and their resulting transcriptomes were compared (47). Eight of the 22 genes described previously by Joseph et al. (47) as being the most highly differentially regulated in MC58 were also differentially regulated in the present study. Seven of the eight genes were similarly upregulated (NMB0879, NMB0995, and NMB1368) or downregulated (NMB0483, NMB1398, NMB1541, and NMB1753) (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). It is noteworthy that NMB0995 was identified in all studies (9, 10, 47) to be upregulated in MC58 in response to cocultivation with epithelial cells.
Considering meningococcal gene transcription during prolonged cocultivation, it is of note that the 16 differentially regulated genes encoding proteins previously identified as being immunoreactive to sera from individuals colonized by meningococci (2) were all upregulated for this period (Table 2), supporting the proposition that our system models growth conditions in vivo during the commensal state. The transcriptional regulators Hfq, PhoP (MisR), and Lrp were upregulated during prolonged (>24 h) cocultivation, establishing their importance in modulating genome-wide patterns of gene expression during established colonization. Hfq binds small RNAs and is widely involved in gene regulation: hfq mutants are attenuated for epithelial cell adherence (48). Employing proteomic approaches, two studies using meningococcal hfq mutants have identified a number of deregulated proteins (49, 50). Genes encoding some deregulated proteins were also found in our differentially regulated gene lists. These genes included pilE, opa, fbpA (NMB0634), metH (NMB0944), glyA (NMB1055), gdhA (NMB1710), groEL (NMB1972), NMB0946, NMB1590, and NMB1796 (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). All were upregulated during prolonged coincubation (24 h, 96 h, and 21 days) but not at 4 h, as was hfq (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). The meningococcal two-component system PhoP/Q regulates a broad range of different genes (51, 52), and a phoP mutant is also attenuated for epithelial cell adherence (53). Meningococcal PhoP-regulated genes have been identified in serogroup B (n = 117) (52) and serogroup C (n = 281) (51). Of these genes, 23 reported by Tzeng et al. (52) and 45 reported by Newcombe et al. (51) were also found to be differentially regulated during prolonged (>24 h) cocultivation (see the data at http://bugs.sgul.ac.uk/E-BUGS-141). Neisserial Lrp is of unknown function, but in other Gram-negative bacteria, it regulates genes involved in various metabolic pathways, including pilus biogenesis (54). Our sustained cocultivation model should assist in delineating the role of these regulators in long-term epithelial cell colonization by meningococci.
In Fig. S6 in the supplemental material, the most enriched category of differentially expressed genes was translation, with most genes being initially downregulated at 4 h but being upregulated during prolonged (>24 h) coincubation with epithelial cells. A similar trend of differential regulation of genes was also observed for those genes categorized in the posttranslational modification, protein turnover, and chaperone categories. This suggests that there is increased protein synthesis activity during prolonged cocultivation. The opposite was observed for meningococci growing in human blood (13), with genes involved in protein synthesis being increasingly downregulated at later cocultivation time points. Thus, there appears to be a similarity in protein synthesis activity in meningococci growing in blood and early (4 h), but not prolonged, cocultivation with epithelial cells.
Using another method of functional grouping of differentially expressed genes (DAVID), the most enriched functional categories were all related to meningococcal surface structures. The most upregulated (FC, 10 to 37) 10 genes at 21 days of cocultivation were those encoding pili (pilE and 5 pilS), opacity proteins (NMB0442 and NMB1465), and two hypothetical proteins (NMB0046 and NMB1116). Their upregulation was not as high at 4 h (FC, 1.65 to 5.1). It has been reported that, compared to 4 h of attachment to epithelial cells, pili are less exposed at the meningococcal surface at 9 h, allowing tight attachment through other adhesins, particularly opacity proteins (55). In contrast, we found upregulation of pilus genes at 21 days, and this correlated with high numbers of adherent meningococci (Fig. 2C; see also Fig. S4 in the supplemental material). It is likely that meningococci constantly colonize new areas of the epithelial monolayer as an adaptation to their environment. It may partly explain why meningococci are able to persist on the nasopharyngeal epithelia of an individual for weeks or months.
We were concerned to demonstrate that 16HBE14 cells in culture remained viable after prolonged coincubation (96 h and 21 days) with meningococci. At 48 h, light microscopic inspection of Giemsa-stained 16HBE14 and COR-L23 cells cocultured with meningococci revealed intact cells associated with many meningococci in the case of the former but dead/dying cells with very few meningococci in the case of the latter (Fig. 1; see also Fig. S4 in the supplemental material). Dead epithelial cells do not associate with meningococci to any substantial extent, a reflection of the fact that association is not nonspecific but receptor mediated (23). We interpret the even higher numbers of meningococci associated with 16HBE14 cells at the later time points to indicate the epithelial cells' continuing viability. A second line of evidence comes from the results of the gentamicin protection assay (see Fig. S4 in the supplemental material). Epithelial cells must be intact and metabolically active to afford protection for intracellular meningococci from externally applied gentamicin.
The five mutants of the NMB0342–NMB0348 locus all showed significant reductions in epithelial cell adherence. As there was no polar effect of any of the gene disruptions studied, the defect in epithelial cell adherence for each mutant was attributable to disruption of the targeted gene. The biological functions of these genes are unknown, except that NMB0345 is homologous to cj0596 from Campylobacter jejuni, encoding a protein known to be important for epithelial cell adherence, colonization of mouse intestinal epithelium, and biofilm formation (56). Further work will be required to ascertain whether specific genes in the NMB0342–NMB0348 locus have a role in outer membrane protein assembly, as was suggested for cj0596 (57), and in promoting epithelial-bacterial cell interactions and/or colonization.
We have proposed previously (13) that meningococcal surface-expressed proteins encoded by genes that are relatively downregulated in the nasopharynx but upregulated in the bloodstream should be comparatively invariant in sequence and attractive candidate vaccine antigens. Three genes found to be upregulated upon coincubation with human blood (NMB0497, with a FC of 1.7; NMB0866, with a FC of 1.2; and NMB1882, with a FC of 2.3) (13) fulfilled this criterion. NMB0497 and NMB0866 were both downregulated at all time points in the present study, while NMB1882 was downregulated at 21 days.
NMB0497, NMB0866, and NMB1882 contain 1, 6, and 12 peptide loops, respectively, predicted to project beyond the meningococcal outer membrane by PRED-TMBB (58), and each loop shows a high degree of sequence conservation between homologues from different meningococcal strains available at the KEGG database (http://www.genome.jp/kegg), compared by using ClustalW. If these proteins are surface located and abundant, they may prove to be useful components to broaden the coverage of existing vaccine formulations, such as the investigational MenB vaccine Bexsero (59), to prevent invasive infections caused by serogroup B strains of N. meningitidis.
Modeling of prolonged meningococcal colonization by epithelial cell cocultivation provides insight into gene expression during this important, but experimentally relatively inaccessible, phase of human infection. Comparison of transcriptomes obtained at early (4 h) and late (24 h, 96 h, and 21 days) cocultivation time points suggests that the regulators PhoP/Q, Hfq, and Lrp have a role in differentially regulating the meningococcal gene repertoire, enabling adaptation at different stages of colonization. Functional grouping showed that transcriptional changes in genes involved in protein synthesis at the early cocultivation time point (4 h) resembled those of meningococci growing in blood (13). This indicates that at an early stage of colonization, bacteria are metabolically adapted for growth in blood. The higher levels of expression of adhesin genes at later time points suggest their importance for prolonged nasopharyngeal epithelial cell colonization.
Supplementary Material
ACKNOWLEDGMENTS
We thank BμG@S (Bacterial Microarray Group at St. George's University of London) for supplying microarrays and advice.
We thank The Wellcome Trust for funding the multicollaborative microbial pathogen microarray facility under its Functional Genomics Resources Initiative. This study was funded by the Health Protection Agency (now Public Health England) and the George John and Sheilah Livanos Charitable Trust.
Footnotes
Published ahead of print 26 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00397-13.
REFERENCES
- 1.Wilder-Smith A, Barkham TM, Ravindran S, Earnest A, Paton NI. 2003. Persistence of W135 Neisseria meningitidis carriage in returning Hajj pilgrims: risk for early and late transmission to household contacts. Emerg. Infect. Dis. 9:123–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams JN, Skipp PJ, O'Connor CD, Christodoulides M, Heckels JE. 2009. Immunoproteomic analysis of the development of natural immunity in subjects colonized by Neisseria meningitidis reveals potential vaccine candidates. Infect. Immun. 77:5080–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davila S, Wright VJ, Khor CC, Sim KS, Binder A, Breunis WB, Inwald D, Nadel S, Betts H, Carrol ED, de Groot R, Hermans PW, Hazelzet J, Emonts M, Lim CC, Kuijpers TW, Martinon-Torres F, Salas A, Zenz W, Levin M, Hibberd ML. 2010. Genome-wide association study identifies variants in the CFH region associated with host susceptibility to meningococcal disease. Nat. Genet. 42:772–776 [DOI] [PubMed] [Google Scholar]
- 4.McCall BJ, Neill AS, Young MM. 2004. Risk factors for invasive meningococcal disease in southern Queensland, 2000-2001. Intern. Med. J. 34:464–468 [DOI] [PubMed] [Google Scholar]
- 5.Seib KL, Pigozzi E, Muzzi A, Gawthorne JA, Delany I, Jennings MP, Rappuoli R. 2011. A novel epigenetic regulator associated with the hypervirulent Neisseria meningitidis clonal complex 41/44. FASEB J. 25:3622–3633 [DOI] [PubMed] [Google Scholar]
- 6.Stephens DS, Greenwood B, Brandtzaeg P. 2007. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369:2196–2210 [DOI] [PubMed] [Google Scholar]
- 7.Neal KR, Nguyen-van Tam JS, Slack RC, Kaczmarski EB, White A, Ala'Aldeen DA. 1999. Seven-week interval between acquisition of a meningococcus and the onset of invasive disease. A case report. Epidemiol. Infect. 123:507–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budroni S, Siena E, Dunning Hotopp JC, Seib KL, Serruto D, Nofroni C, Comanducci M, Riley DR, Daugherty SC, Angiuoli SV, Covacci A, Pizza M, Rappuoli R, Moxon ER, Tettelin H, Medini D. 2011. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 108:4494–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietrich G, Kurz S, Hubner C, Aepinus C, Theiss S, Guckenberger M, Panzner U, Weber J, Frosch M. 2003. Transcriptome analysis of Neisseria meningitidis during infection. J. Bacteriol. 185:155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grifantini R, Bartolini E, Muzzi A, Draghi M, Frigimelica E, Berger J, Ratti G, Petracca R, Galli G, Agnusdei M, Giuliani MM, Santini L, Brunelli B, Tettelin H, Rappuoli R, Randazzo F, Grandi G. 2002. Previously unrecognized vaccine candidates against group B meningococcus identified by DNA microarrays. Nat. Biotechnol. 20:914–921 [DOI] [PubMed] [Google Scholar]
- 11.Gruenert DC, Finkbeiner WE, Widdicombe JH. 1995. Culture and transformation of human airway epithelial cells. Am. J. Physiol. 268:L347–L360 http://ajplung.physiology.org/content/268/3/L347.abstract [DOI] [PubMed] [Google Scholar]
- 12.O'Dwyer CA, Li MS, Langford PR, Kroll JS. 2009. Meningococcal biofilm growth on an abiotic surface—a model for epithelial colonization? Microbiology 155:1940–1952 [DOI] [PubMed] [Google Scholar]
- 13.Hedman AK, Li MS, Langford PR, Kroll JS. 2012. Transcriptional profiling of serogroup B Neisseria meningitidis growing in human blood: an approach to vaccine antigen discovery. PLoS One 7:e39718. 10.1371/journal.pone.0039718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4:P3. 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-delta delta C(T) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 16.Rusniok C, Vallenet D, Floquet S, Ewles H, Mouze-Soulama C, Brown D, Lajus A, Buchrieser C, Medigue C, Glaser P, Pelicic V. 2009. NeMeSys: a biological resource for narrowing the gap between sequence and function in the human pathogen Neisseria meningitidis. Genome Biol. 10:R110. 10.1186/gb-2009-10-10-r110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, Ketchum KA, Hood DW, Peden JF, Dodson RJ, Nelson WC, Gwinn ML, DeBoy R, Peterson JD, Hickey EK, Haft DH, Salzberg SL, White O, Fleischmann RD, Dougherty BA, Mason T, Ciecko A, Parksey DS, Blair E, Cittone H, Clark EB, Cotton MD, Utterback TR, Khouri H, Qin H, Vamathevan J, Gill J, Scarlato V, Masignani V, Pizza M, Grandi G, Sun L, Smith HO, Fraser CM, Moxon ER, Rappuoli R, Venter JC. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809–1815 [DOI] [PubMed] [Google Scholar]
- 18.Pelicic V, Morelle S, Lampe D, Nassif X. 2000. Mutagenesis of Neisseria meningitidis by in vitro transposition of Himar1 mariner. J. Bacteriol. 182:5391–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tzeng YL, Ambrose KD, Zughaier S, Zhou X, Miller YK, Shafer WM, Stephens DS. 2005. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J. Bacteriol. 187:5387–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray D. 1992. Cell movements. Garland Publishing Co., New York, NY [Google Scholar]
- 21.DeMali KA, Burridge K. 2003. Coupling membrane protrusion and cell adhesion. J. Cell Sci. 116:2389–2397 [DOI] [PubMed] [Google Scholar]
- 22.Mikaty G, Soyer M, Mairey E, Henry N, Dyer D, Forest KT, Morand P, Guadagnini S, Prevost MC, Nassif X, Dumenil G. 2009. Extracellular bacterial pathogen induces host cell surface reorganization to resist shear stress. PLoS Pathog. 5:e1000314. 10.1371/journal.ppat.1000314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carbonnelle E, Hill DJ, Morand P, Griffiths NJ, Bourdoulous S, Murillo I, Nassif X, Virji M. 2009. Meningococcal interactions with the host. Vaccine 27(Suppl 2):B78–B89. 10.1016/j.vaccine.2009.04.069 [DOI] [PubMed] [Google Scholar]
- 24.Brown DR, Helaine S, Carbonnelle E, Pelicic V. 2010. Systematic functional analysis reveals that a set of seven genes is involved in fine-tuning of the multiple functions mediated by type IV pili in Neisseria meningitidis. Infect. Immun. 78:3053–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carbonnelle E, Helaine S, Prouvensier L, Nassif X, Pelicic V. 2005. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol. Microbiol. 55:54–64 [DOI] [PubMed] [Google Scholar]
- 26.Virji M, Makepeace K, Ferguson DJ, Achtman M, Moxon ER. 1993. Meningococcal Opa and Opc proteins: their role in colonization and invasion of human epithelial and endothelial cells. Mol. Microbiol. 10:499–510 [DOI] [PubMed] [Google Scholar]
- 27.Monaco C, Tala A, Spinosa MR, Progida C, De Nitto E, Gaballo A, Bruni CB, Bucci C, Alifano P. 2006. Identification of a meningococcal L-glutamate ABC transporter operon essential for growth in low-sodium environments. Infect. Immun. 74:1725–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi H, Kim KS, Watanabe H. 2011. Meningococcal internalization into human endothelial and epithelial cells is triggered by the influx of extracellular L-glutamate via GltT L-glutamate ABC transporter in Neisseria meningitidis. Infect. Immun. 79:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar P, Sannigrahi S, Tzeng YL. 2012. The Neisseria meningitidis ZnuD zinc receptor contributes to interactions with epithelial cells and supports heme utilization when expressed in Escherichia coli. Infect. Immun. 80:657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammerschmidt S, Hilse R, van Putten JP, Gerardy-Schahn R, Unkmeir A, Frosch M. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 15:192–198 [PMC free article] [PubMed] [Google Scholar]
- 31.Kahler CM, Martin LE, Shih GC, Rahman MM, Carlson RW, Stephens DS. 1998. The (alpha2→8)-linked polysialic acid capsule and lipooligosaccharide structure both contribute to the ability of serogroup B Neisseria meningitidis to resist the bactericidal activity of normal human serum. Infect. Immun. 66:5939–5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plant L, Sundqvist J, Zughaier S, Lovkvist L, Stephens DS, Jonsson AB. 2006. Lipooligosaccharide structure contributes to multiple steps in the virulence of Neisseria meningitidis. Infect. Immun. 74:1360–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi H, Carlson RW, Muszynski A, Choudhury B, Kim KS, Stephens DS, Watanabe H. 2008. Modification of lipooligosaccharide with phosphoethanolamine by LptA in Neisseria meningitidis enhances meningococcal adhesion to human endothelial and epithelial cells. Infect. Immun. 76:5777–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bos MP, Tommassen J. 2011. The LptD chaperone LptE is not directly involved in lipopolysaccharide transport in Neisseria meningitidis. J. Biol. Chem. 286:28688–28696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakshi S. 2004. Role of the NMB0342-NMB0348 locus in meningococcal pathogenesis and investigation of NMB0345 as a vaccine candidate. Ph.D. thesis University of Oxford, Oxford, United Kingdom [Google Scholar]
- 36.Sun YH, Bakshi S, Chalmers R, Tang CM. 2000. Functional genomics of Neisseria meningitidis pathogenesis. Nat. Med. 6:1269–1273 [DOI] [PubMed] [Google Scholar]
- 37.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersson A, Kortekaas J, Weynants VE, Voet P, Poolman JT, Bos MP, Tommassen J. 2006. Vaccine potential of the Neisseria meningitidis lactoferrin-binding proteins LbpA and LbpB. Vaccine 24:3545–3557 [DOI] [PubMed] [Google Scholar]
- 39.Rokbi B, Renauld-Mongenie G, Mignon M, Danve B, Poncet D, Chabanel C, Caugant DA, Quentin-Millet MJ. 2000. Allelic diversity of the two transferrin binding protein B gene isotypes among a collection of Neisseria meningitidis strains representative of serogroup B disease: implication for the composition of a recombinant TbpB-based vaccine. Infect. Immun. 68:4938–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West D, Reddin K, Matheson M, Heath R, Funnell S, Hudson M, Robinson A, Gorringe A. 2001. Recombinant Neisseria meningitidis transferrin binding protein A protects against experimental meningococcal infection. Infect. Immun. 69:1561–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marzoa J, Sanchez S, Costoya L, Dieguez-Casal E, Freixeiro P, Brookes C, Allen L, Taylor S, Gorringe AR, Ferreiros CM, Criado MT. 2012. Induction of immune responses by purified outer membrane protein complexes from Neisseria meningitidis. Vaccine 30:2387–2395 [DOI] [PubMed] [Google Scholar]
- 42.Turner DP, Marietou AG, Johnston L, Ho KK, Rogers AJ, Wooldridge KG, Ala'Aldeen DA. 2006. Characterization of MspA, an immunogenic autotransporter protein that mediates adhesion to epithelial and endothelial cells in Neisseria meningitidis. Infect. Immun. 74:2957–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner DP, Wooldridge KG, Ala'Aldeen DA. 2002. Autotransported serine protease A of Neisseria meningitidis: an immunogenic, surface-exposed outer membrane, and secreted protein. Infect. Immun. 70:4447–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendum TA, Newcombe J, McNeilly CL, McFadden J. 2009. Towards the immunoproteome of Neisseria meningitidis. PLoS One 4:e5940. 10.1371/journal.pone.0005940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serruto D, Bottomley MJ, Ram S, Giuliani MM, Rappuoli R. 2012. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: immunological, functional and structural characterization of the antigens. Vaccine 30(Suppl 2):B87–B97. 10.1016/j.vaccine.2012.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuwae A, Sjolinder H, Eriksson J, Eriksson S, Chen Y, Jonsson AB. 2011. NafA negatively controls Neisseria meningitidis piliation. PLoS One 6:e21749. 10.1371/journal.pone.0021749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joseph B, Schneiker-Bekel S, Schramm-Gluck A, Blom J, Claus H, Linke B, Schwarz RF, Becker A, Goesmann A, Frosch M, Schoen C. 2010. Comparative genome biology of a serogroup B carriage and disease strain supports a polygenic nature of meningococcal virulence. J. Bacteriol. 192:5363–5377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dietrich M, Munke R, Gottschald M, Ziska E, Boettcher JP, Mollenkopf H, Friedrich A. 2009. The effect of hfq on global gene expression and virulence in Neisseria gonorrhoeae. FEBS J. 276:5507–5520 [DOI] [PubMed] [Google Scholar]
- 49.Fantappie L, Metruccio MM, Seib KL, Oriente F, Cartocci E, Ferlicca F, Giuliani MM, Scarlato V, Delany I. 2009. The RNA chaperone Hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect. Immun. 77:1842–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pannekoek Y, Huis in't Veld R, Hopman CT, Langerak AA, Speijer D, van der Ende A. 2009. Molecular characterization and identification of proteins regulated by Hfq in Neisseria meningitidis. FEMS Microbiol. Lett. 294:216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newcombe J, Jeynes JC, Mendoza E, Hinds J, Marsden GL, Stabler RA, Marti M, McFadden JJ. 2005. Phenotypic and transcriptional characterization of the meningococcal PhoPQ system, a magnesium-sensing two-component regulatory system that controls genes involved in remodeling the meningococcal cell surface. J. Bacteriol. 187:4967–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tzeng YL, Kahler CM, Zhang X, Stephens DS. 2008. MisR/MisS two-component regulon in Neisseria meningitidis. Infect. Immun. 76:704–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson CR, Newcombe J, Thorne S, Borde HA, Eales-Reynolds LJ, Gorringe AR, Funnell SG, McFadden JJ. 2001. Generation and characterization of a PhoP homologue mutant of Neisseria meningitidis. Mol. Microbiol. 39:1345–1355 [DOI] [PubMed] [Google Scholar]
- 54.Calvo JM, Matthews RG. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58:466–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pujol C, Eugene E, Marceau M, Nassif X. 1999. The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc. Natl. Acad. Sci. U. S. A. 96:4017–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Asakura H, Yamasaki M, Yamamoto S, Igimi S. 2007. Deletion of peb4 gene impairs cell adhesion and biofilm formation in Campylobacter jejuni. FEMS Microbiol. Lett. 275:278–285 [DOI] [PubMed] [Google Scholar]
- 57.Kale A, Phansopa C, Suwannachart C, Craven CJ, Rafferty JB, Kelly DJ. 2011. The virulence factor PEB4 (Cj0596) and the periplasmic protein Cj1289 are two structurally related SurA-like chaperones in the human pathogen Campylobacter jejuni. J. Biol. Chem. 286:21254–21265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. 2004. PRED-TMBB: a Web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 32:W400–W404. 10.1093/nar/gkh417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorringe AR, Pajon R. 2012. Bexsero: a multicomponent vaccine for prevention of meningococcal disease. Hum. Vaccin. Immunother. 8:174–183 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


