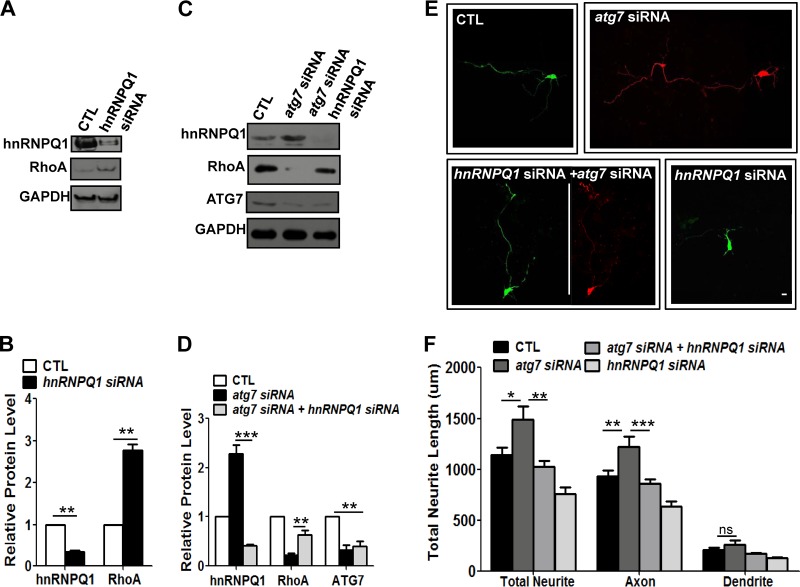
Fig 8.
hnRNP-Q1 regulated by autophagy negatively regulates RhoA and affects early axon growth in cultured neurons. (A and B) Western blot (A) and quantification plots of bands (B) of cultured cortical neurons at DIV1 to DIV3 transfected with hnRNP-Q1 siRNA showed a significant decrease in hnRNP-Q1 levels (B) and a significant increase in RhoA levels (B) compared to the control (scrambled siRNA), indicating that hnRNP-Q1 negatively regulates RhoA in cortical neurons. GAPDH was used as a loading control and a normalizing band for quantification. The values are the means and SEM of three independent replicates. Student's t test; **, P < 0.01. (C and D) Western blot (C) and quantification plots of bands (D) of cultured cortical neurons at DIV2 and DIV3 with cotransfection of atg7 and hnRNP-Q1 siRNA showing recovery of the autophagy deficiency-mediated reduction in RhoA levels. GAPDH was used as a loading control. The values are the means and SEM of three independent replicates. One-way ANOVA and Tukey's multiple-comparison test; **, P < 0.01; ***, P < 0.001. (E and F) Composite images of neurite morphology (E) were generated from 2 or 3 separate images obtained from the same neuron to track the total neurite length of atg7 siRNA-transfected neurons. Representative images of neurite growth (E) and a quantification plot of the neurite length (F) of cultured cortical neurons at DIV1 to DIV3 transfected with atg7 siRNA, hnRNP-Q1 siRNA, or both show that cotransfection of atg7 siRNA (in pSICOR-RFP) with hnRNP-Q1 siRNA (in pSUPER-GFP) significantly recued the axon overextension phenotype, indicating that reduction of hnRNP-Q1 recovers the level of RhoA and suppresses abnormal axon elongation. Transfection with hnRNP-Q1 siRNA alone showed a slight reduction in neurite length compared to the control. Transfection with hnRNP-Q1 or atg7 siRNA and neurite morphology was determined by expression of GFP or RFP, respectively, or both. CTL, control neurons transfected with scrambled siRNA. The values are shown as the means and SEM of three independent replicates. One-way ANOVA and Tukey's multiple-comparison test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant. Scale bar, 20 μm.