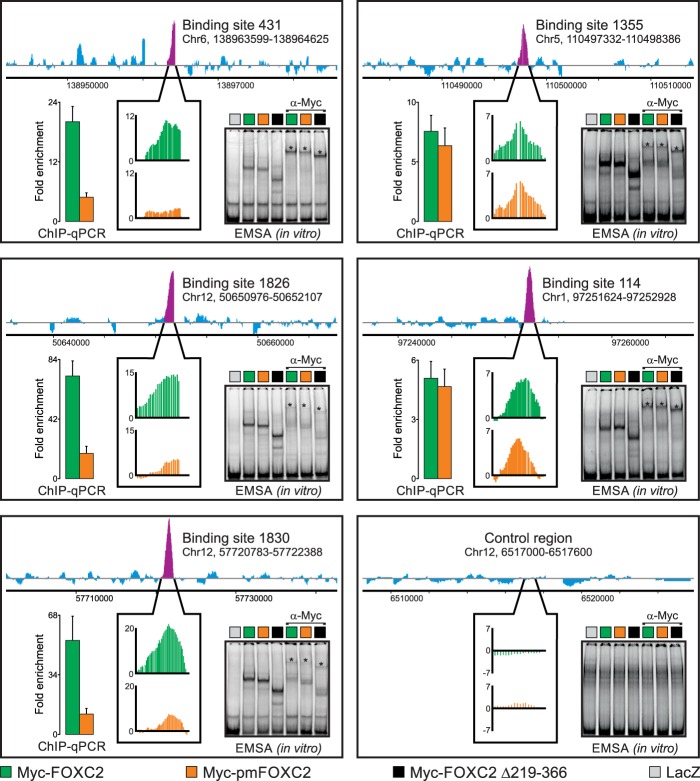
Fig 4.
Phosphorylation differentially regulates FOXC2 binding to genomic target sites in the context of native chromatin but not in vitro. We used genome-wide ChIP-chip to compare the binding of adenovirus-expressed wild-type Myc-FOXC2 and the phosphorylation-deficient mutant Myc-pmFOXC2 to physiological binding sites occupied by endogenous FOXC2 in primary LECs. Endogenous FOXC2 enrichment profiles are shown at the top of each panel. Purple peaks indicate FOXC2-enriched regions; their relative occupancies by Myc-FOXC2 and Myc-pmFOXC2 are shown in callout boxes in green and orange, respectively. Vertical axes represent MAT score. Binding sites are numbered as in Norrmén et al. (8); genomic coordinates refer to the hg18 human genome assembly. An unbound control region is shown in the lower right panel. The ChIP-chip results were validated by ChIP-qPCR with primers flanking ∼100-bp sequences within the FOXC2-enriched regions. The results are presented as the fold enrichment over the unbound control region. Green and orange bars correspond to wild-type Myc-FOXC2 and Myc-pmFOXC2, respectively. Shown are the means and standard deviations of triplicate determinations. An EMSA was used to compare the in vitro binding of adenovirus-expressed wild-type Myc-FOXC2, Myc-pmFOXC2, and deletion mutant Myc-FOXC2 D219-366 to naked dsDNA from the ChIP-enriched regions or the unbound control region. Binding specificity was controlled with adenovirus-expressed bacterial β-galactosidase (LacZ) and anti-Myc antibody. Asterisks indicate the positions of the antibody-supershifted complexes.