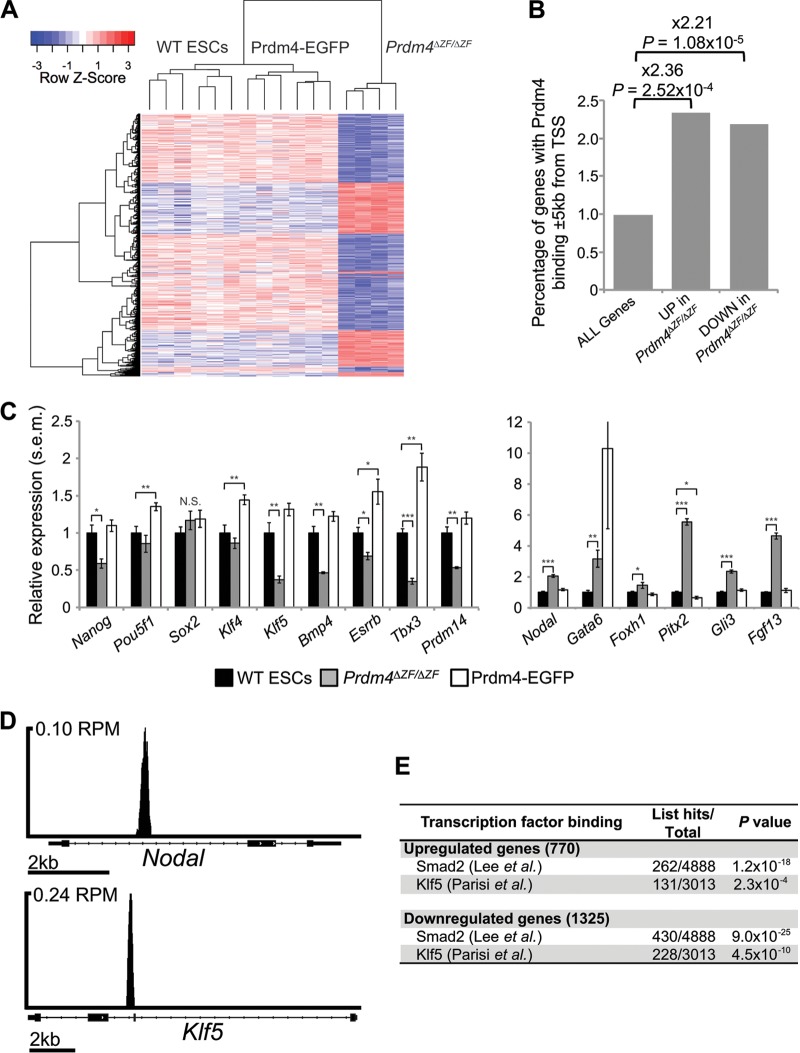
Fig 7.
Prdm4 functional loss causes misregulated expression of pluripotency/differentiation genes, including genes with proximal Prdm4 binding. (A) Hierarchical clustering of microarray data reveals distinct transcriptional signatures between Prdm4ΔZF/ΔZF ESCs and wild-type and stably transfected cells expressing Prdm4-EGFP. Represented are the 2,095 genes differentially expressed between Prdm4ΔZF/ΔZF and wild-type ESCs (vertical) across the 16 independent samples (horizontal). The heat map represents standard score (the number of standard deviations removed from the average intensity per row). Samples with lower than the average intensity are blue; those with higher than the average intensity are red. (B) Genes either up- or downregulated in Prdm4ΔZF/ΔZF cells are significantly more likely to have Prdm4 binding within 5 kb of their TSS than are all genes. (C) qPCR shows that pluripotency markers are significantly downregulated in Prdm4ΔZF/ΔZF cells and upregulated in Prdm4-EGFP-expressing cells (left) whereas differentiation markers are upregulated in Prdm4ΔZF/ΔZF cells (right). *, P < 1 × 10−2; **, P < 1 × 10−3; ***, P < 1 × 10−5; N.S., not significant. (D) The TGF-β gene family member Nodal and pluripotency factor gene Klf5 show proximal binding of Prdm4; TSSs are to the left. RPM, peak height in reads per million. (E) Up- and downregulated genes are significantly more likely to have Smad2 binding and Klf5 binding within 10 kb of the TSSs.