Fig 7.
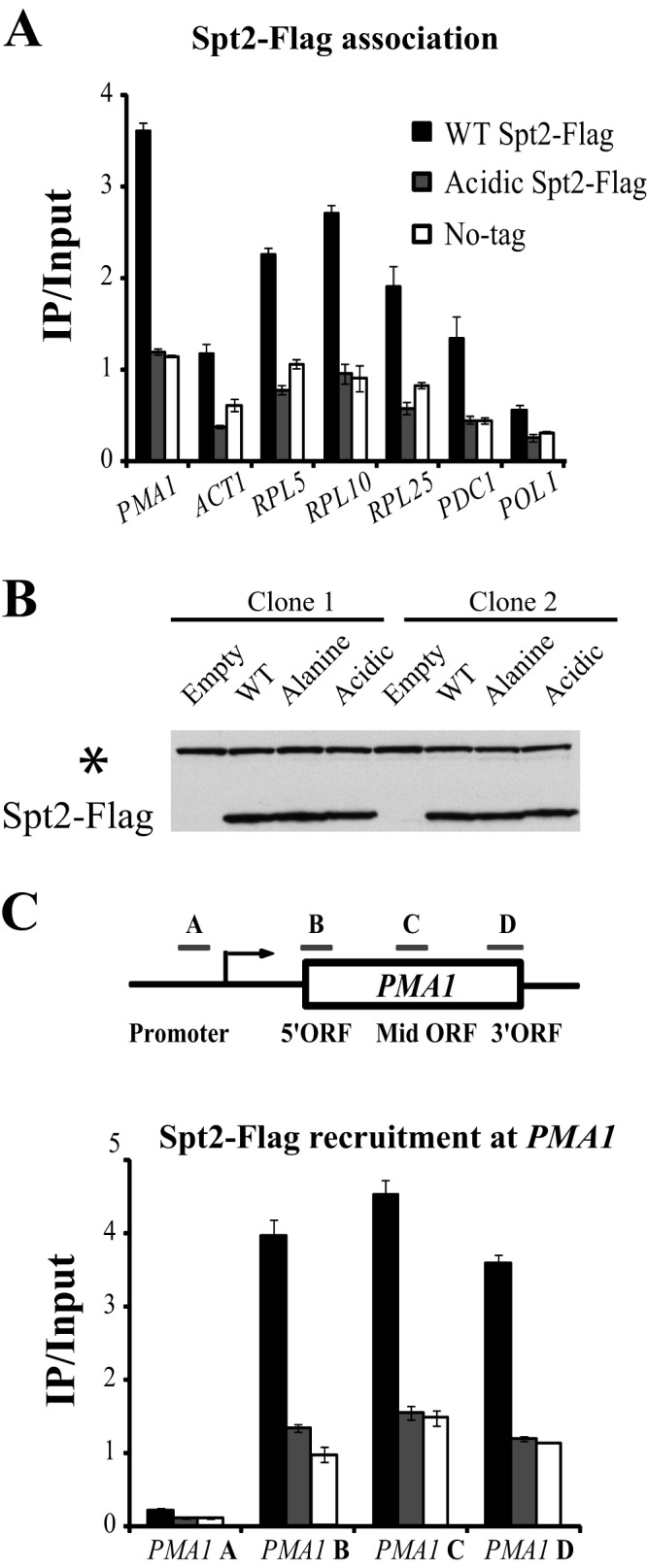
Mimicking CK2 phosphorylation results in the loss of Spt2 from the transcribed regions of different genes. (A) Mutation of Spt2 RI/II to acidic residues results in a dramatic decrease of the Spt2-Flag association with coding regions of different genes. Yeast cells from spt2Δ strains containing a plasmid expressing Flag-tagged wild-type Spt2 or the RI/II acidic version of Spt2 were grown in yeast extract-peptone-dextrose to mid-log phase prior to formaldehyde treatment. Chromatin immunoprecipitations were then performed by using the anti-Flag antibody. The values shown (IP [immunoprecipitated]/Input) represent the averages and standard errors of two to three independent experiments. For each gene, one region located generally near the middle of the open reading frame was tested. (B) Mutation in RI/II sites does not affect the protein level of Spt2 in yeast cells. Yeast cells from the spt2Δ strains were transformed with an empty vector or a plasmid encoding wild-type, acidic, or nonphosphorylatable Spt2 fused to a Flag tag. The WCEs of these cells were subjected to Western blotting with anti-Flag antibodies. The asterisk indicates a protein that was recognized nonspecifically by the antibody and could be used as a loading control. (C) The spt2 mutant mimicking CK2 phosphorylation is lost from all transcribed regions of the active gene PMA1. The chromatin used in this experiment was prepared from the same strains as those in panel A and treated as described above for panel A. The bars labeled A to D represent the PMA1 regions assayed by quantitative PCR. The values shown (IP/Input) represent the averages and standard errors of two to three independent experiments.
