Figure 2. The workflow of data processing.
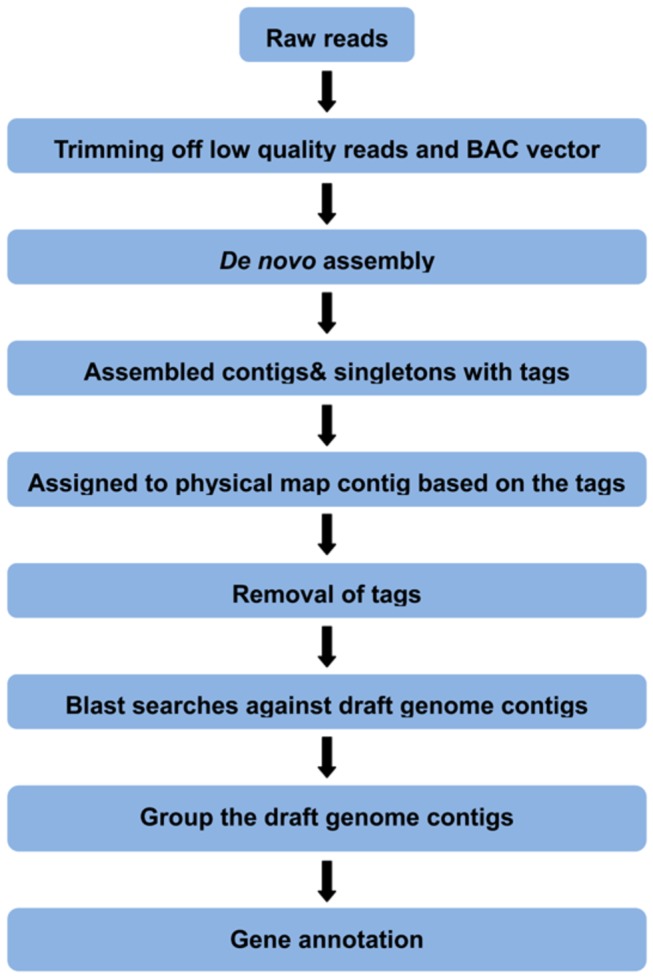
The raw reads were first trimmed off the low quality reads (Q20) and BAC vectors. De novo assembly was then conducted with the filtered high quality reads. The assembled contigs with tag on it, plus singletons which have tag on it were then assigned to each physical contig, based on the specific tag. The tags were removed. The clean sequences were then used as queries to BLAST search against the draft catfish whole genome contigs. The targeted genome contigs were then retrieved and annotated.
