Figure 2. Comparing experimental and calculated relative binding free energies.
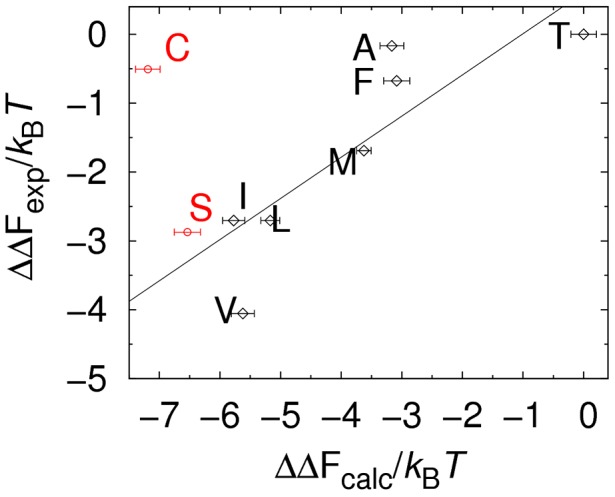
As a quantitative test of the all-atom model [36] used in this work, we calculate binding free energies, 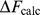 , for the protein PSD95 and 9 different peptide sequences. The
, for the protein PSD95 and 9 different peptide sequences. The  values were obtained from protein-peptide binding simulations performed separately for each PSD95-peptide pair according to our previous protocol [35], [36] (see also Methods) and using Equation 2. The peptide sequences considered are derived from KKETEV (black diamond) and are either
values were obtained from protein-peptide binding simulations performed separately for each PSD95-peptide pair according to our previous protocol [35], [36] (see also Methods) and using Equation 2. The peptide sequences considered are derived from KKETEV (black diamond) and are either  -variants, KKETE-[I/L/M/F/A/T], (black diamonds) or
-variants, KKETE-[I/L/M/F/A/T], (black diamonds) or 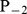 -variants, KKE[S/C]EV (red circles). All simulations were performed at
-variants, KKE[S/C]EV (red circles). All simulations were performed at 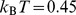 and standard errors were estimated from 10 independent runs. Experimental binding free energies,
and standard errors were estimated from 10 independent runs. Experimental binding free energies, 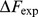 , are taken from Ref. [46]. Both
, are taken from Ref. [46]. Both 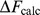 and
and 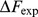 values are shown relative to the weakest binding peptide. The solid line represents the best linear fit, exluding KKECEV, and the correlation coefficient is
values are shown relative to the weakest binding peptide. The solid line represents the best linear fit, exluding KKECEV, and the correlation coefficient is 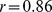 .
.
