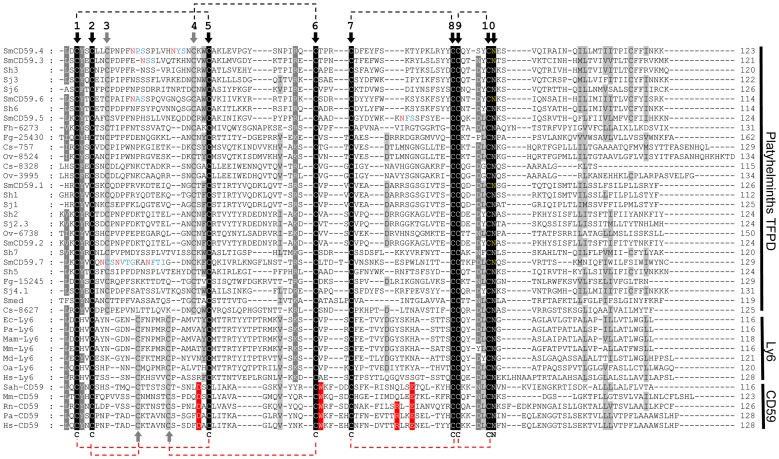
Figure 1. ClustalX multiple sequence alignment of the mature protein sequence (excluding the signal peptide) of TFPDs from platyhelminthes, CD59 and Ly6.
The regions with high identity and similarity between sequences are shown as black and gray columns, according to the ClustalX algorithm. Arrows indicate highly conserved Cysteines and Asparagines with a C and an N, respectively. Dashed lines represent pairs of cysteine residues forming disulfide bonds determined from Hs-CD59 (red) and predicted for SmCD59.2 (black). Only for SmCD59 sequences, potential sites for N-glycosylation are shown in blue with asparagine (N) in red, and potential sites for GPI anchor are shown in yellow. Human CD59 active sites are shaded in red. The sequences abbreviation are: Schistosoma mansoni (SmCD59.1-7), Schistosoma japonicum (Sj1, Sj2.3, Sj3, Sj4.1, Sj6), Schistosoma hematobium (Sh1-3 and Sh5-7), Clonorchis sinensis (Cs-757, Cs-8328, Cs-8627), Opisthorchis viverrini (Ov-8524, Ov-3995 and Ov-6738), Fasciola hepatica (Fh-6273), Fasciola gigantica (Fg-25430 and Fg-15245), Schmidtea mediterranea (Smed), Equus caballus (Ec-Ly6), Pongo abelii (Pa-Ly6 and Pa-CD59), Macaca mulatta (Mam-Ly6), Mus musculus (Mm-Ly6 and Mm-CD59), Monodelphis domestica (Md-Ly6), Ornithorhynchus anatinus (Oa-Ly6), Homo sapiens (Hs-Ly6 and Hs-CD59), Saimiriine herpesvirus (Sah-CD59), Rattus norvegicus (Rn-CD59) (the accession numbers are listed in the Table S2).