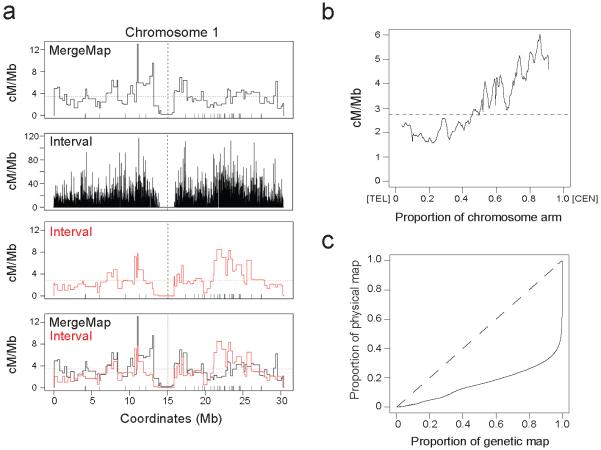
Figure 1. Meiotic crossover frequency in the Arabidopsis genome.
(a) Plots show crossover frequency (cM/Mb) along chromosome 1 estimated by MergeMap (black), Interval (black), Interval plotted using MergeMap markers (red) and the overlay of MergeMap and Interval maps. MergeMap was generated previously from analysis of genotype data from 17 × F populations33 using the MergeMap program35,84. The ‘bin’ widths are variable because they are determined by intermarker distances. Horizontal dashed lines represent mean crossover frequency, vertical dashed lines represent centromeres and vertical ticks above the x-axis indicate disease resistance gene (R-gene) positions. (b) Mean crossover frequency (cM/Mb) estimated by Interval as a proportion along the length of the chromosome arms, orientated with the telomere (TEL) at 0 on the x-axis and the centromere (CEN) at 1. (c) The proportion of the crossovers estimated by Interval plotted against the proportion of physical sequence (solid black line). The dashed line represents a uniform relationship between the genetic and physical maps.