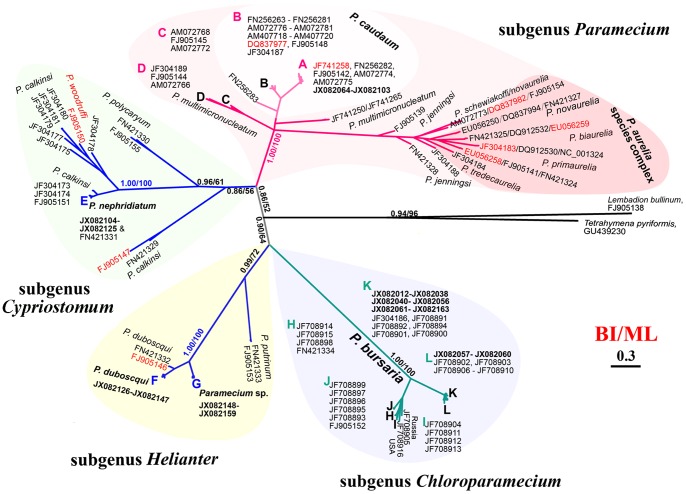
Figure 4. Phylogenetic tree of the barcoding region of 263 cytochrome c oxidase subunit I (COI) gene sequences of the genus Paramecium and genera Lembadion and, Tetrahymena inferred by Bayesian Inference (BI) analysis based on dataset COI-f.
The branches are shaded according to subgenera Chloroparamecium, Helianter, Cypriostomum, Paramecium, proposed by Fokin et al. [28]. The scale bar corresponds to 30 substitutions per 100 nucleotide positions. For P. bursaria, Clade H includes populations sampled from Australia, Germany, and Poland; Clade I and J include populations sampled from Russia and Poland, Germany, Ukraine, and Canada; Clade K includes populations sampled from China (Pb1C1-4 & Pb2C &Pb3C2-3), Austria, Japan, and Italy; Clade L includes populations sampled from China (Pb3C1), Russia, and Japan (see details in Fig. S2 in file S3). For P.caudatum, Clade A includes populations sampled from China (PcC1-4 and AM072774), Australia, USA, and Brazil while members of Clade B were sampled from Germany, Italy, Russia, UK, Norway, Hungary, Slovenia, and Austria (see details in Fig. S3 in file S3). Inconsistent sequences (FJ905146, FJ905147, EU056259, EU056258, DQ837977, DQ837982, JF741258, JF304183) are marked in red [14], [42], [47].