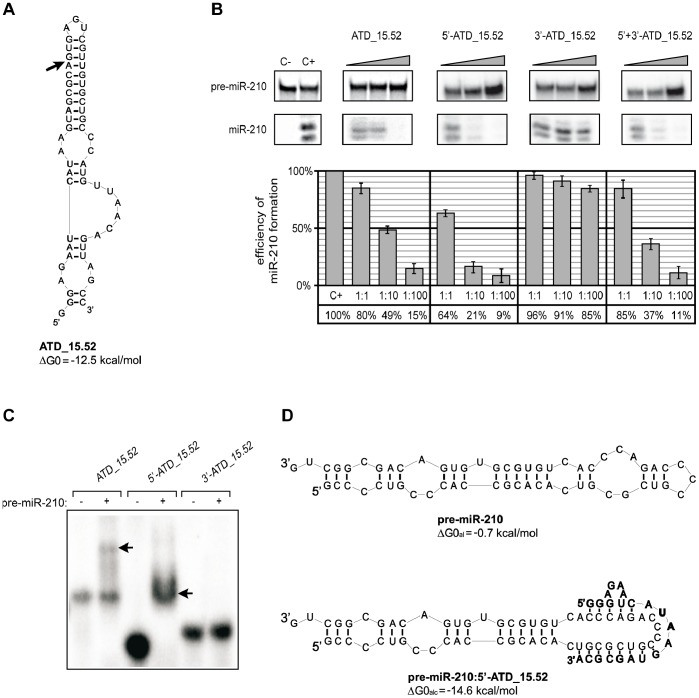
Figure 2. ATD_15.52 is cut by hDicer, and the cleavage product functions as a specific inhibitor.
(A) Predicted secondary structure of ATD_15.52. The ΔG0 value, expressed in kcal/mol, is shown next to the structure. The arrow indicates the determined hDicer cleavage site. (B) Influence of ATD_15.52 and its two hDicer cleavage fragments on the efficiency of miR-210 production. Radiolabeled pre-miR-210 was incubated with hDicer in the presence of ATD_15.52, one of its fragments (5′- or 3′-ATD_15.52) or both fragments (5′+3′ ATD_15.52). Control reactions lacked the enzyme and oligomers (C−) or oligomers only (C+). Triangles indicate increasing concentrations of applied oligomers (pre-miRNA:oligomer molar ratios of 1∶1, 1∶10, and 1∶100). The diagrams show the average efficiency of miR-210 production based on three independent experiments; error bars represent the standard deviations. (C) Binding of ATD_15.52 and its fragments to pre-miR-210. Radiolabeled ATD_15.52 or one of its fragments (5′-ATD_15.52 or 3′-ATD_15.52) was denatured and renatured alone (−) or in the presence of pre-miR-210 (+). The reactions were separated in a native polyacrylamide gel. The positions of the complexes formed by ATD_15.52 or its 5′-fragment and pre-miR-210 are indicated by arrows. (D) The predicted secondary structures of pre-miR-210 (regular) and its complex with the 5′ fragment of ATD_15.52 (bold). The free energies calculated for the apical fragment of pre-miR-210 (ΔG0al) or for the complex formed by the corresponding apical fragment of pre-miR-210 and 5′-ATD_15.52 (ΔG0alc) are shown next to the structures.