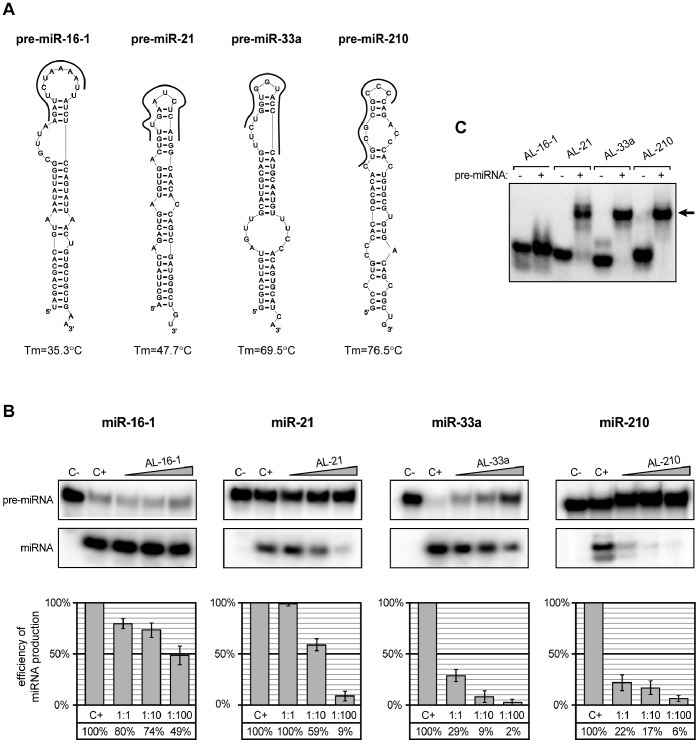
Figure 3. Twelve-nucleotide oligomers targeting apical fragments of pre-miRNA hairpins disturb precursor processing by hDicer.
(A) The predicted secondary structures of four tested pre-miRNAs (pre-miR-16-1, pre-miR-21, pre-miR-33a and pre-miR-210). Oligonucleotides are shown as thick, black curves. The calculated melting temperatures of duplexes formed by nucleotides present in the apical fragments of each pre-miRNA and corresponding oligomer are shown below the structures. (B) Radiolabeled pre-miRNAs were incubated with hDicer in the presence of the appropriate 12-nt oligomers. Control reactions lacked the enzyme and oligomer (C−) or the oligomer only (C+). Triangles represent increasing amounts of indicated oligomers (pre-miRNA:oligomer molar ratios of 1∶1, 1∶10, and 1∶100). The diagrams show the average efficiency of miRNA production based on three independent experiments; error bars represent the standard deviations. (C) Binding of the 12-nt oligomers to the corresponding pre-miRNAs. Radiolabeled oligomers were denatured and renatured alone (−) or in the presence of the corresponding pre-miRNAs (+). Reactions were separated in a native polyacrylamide gel. The position of complexes formed by oligomers and pre-miRNAs are indicated with arrows.