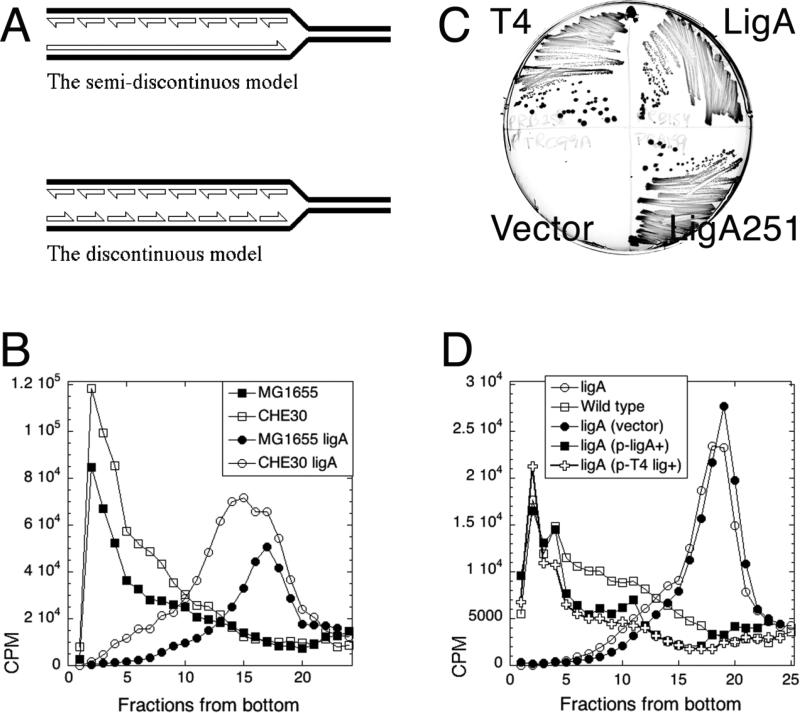
Fig. 1. The test of two models of DNA replication in the ligase mutant: background-independence and complementation of the ligase defect.
A. The two models of intermediates in DNA replication. Half-arrowheads, 3'-ends. The LMW intermediates (Okazaki fragments) are eventually linked together to form mature DNA strands. B. LMW intermediates are background-independent. Alkaline 5-20% sucrose gradients were run from right to left and collected from bottom. In this and all subsequent gradients, fractions 1-5 are considered high molecular weight (HMW), whereas fractions 15 and up are low molecular weight (LMW). Strains are: CHE30, GR523; MG1655 ligA, LA98; CHE30 ligA, GR501. C. Phenotypic complementation. MG1655 ligA251 (LA98) was transformed with plasmids expressing either T4 DNA ligase, or WT E. coli ligase, or the LigA251 mutant protein, as well as with the control vector. Strains were streaked on to agar plates and incubated overnight at 42°C. D. Complementation with functional ligase genes from plasmids. Strains are: ligA, LA98; wild type, MG1655. Plasmids are: vector, pTRC99a; p-ligA+, pRB154; p-T4 lig+, pRB258.