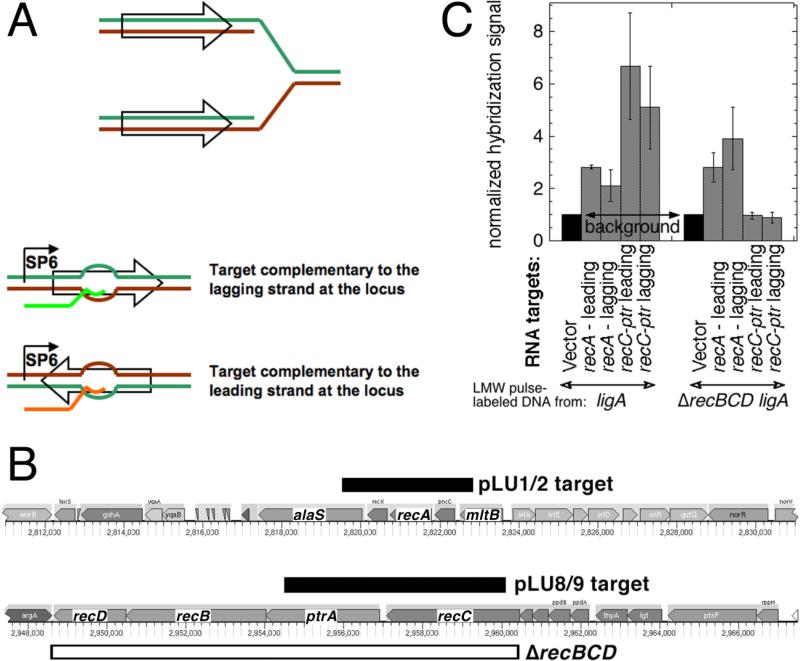
Fig. 4. LMW replication intermediates hybridize to both DNA strands.
A. Strand-specific RNA targets were generated by in vitro SP6 polymerase transcription of chromosomal fragments (shown as empty arrows on a replication fork) cloned under the SP6 promoter in a vector (pSP72) and deposited on a hybridization membrane. The “Watson” strand at the chromosomal locus is labeled green, while the “Crick” strand is labeled brown. The generated RNA target identical to the Watson strand is shown in bright green, while the one identical to the Crick strand is shown in orange. B. The chromosomal position and the actual length of the fragments (pLU1/2 and pLU8/9 targets) is shown as black rectangles. The span of the ΔrecBCD deletion in the AK148 strain, used for the negative control, is shown as the open rectangle. The direction of replication for both regions is from right to left. C. Hybridization of pulsed-labeled LMW DNA to strand-specific RNA targets at the two chromosomal loci. Transcripts from vector-only were deposited to generate the background reading. 3H-labeled replication intermediates from ligA (GR501) or ligA ΔrecBCD (AK148) mutant cells were isolated by alkaline sucrose gradients and used as probes for the membranes with the deposited strand-specific targets. The total counts from various targets in any given experiment were normalized to the vector counts. The values are averages of 4-6 independent measurements ± SEM.