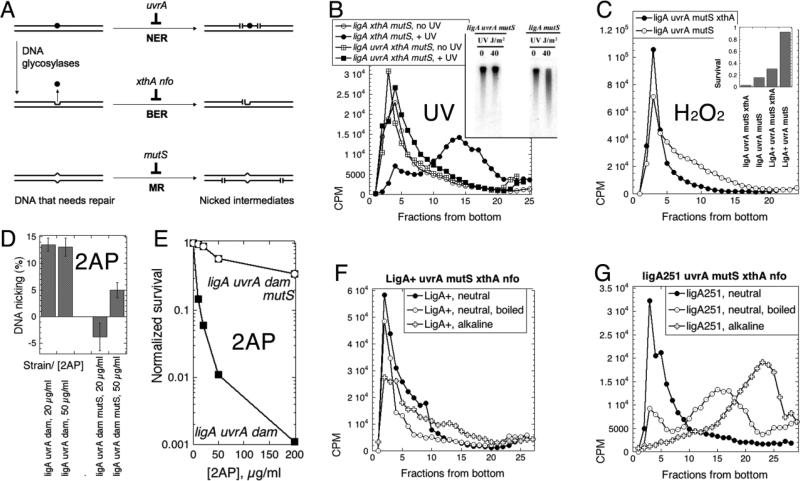
Fig. 5. Inactivation of the major DNA-scission activities does not affect the LMW replication intermediates in the ligA251 mutant.
Panels B-E are labeled with the DNA-damaging agents tested. A. A scheme of the major DNA repair pathways going through strand scission intermediates. DNA duplex is shown as a double line; DNA lesion is indicated by black circle; DNA mismatch is the bump in the duplex; abasic site is the indent in one of the DNA strands, left after the base was removed. NER, nucleotide excision repair; BER, base excision repair; MR, mismatch removal. The scenario shown (that the same DNA lesion is the substrate for both NER and BER) is not a common one and is shown for parsimony: typical DNA lesions are substrates to either NER or BER, but not both. B. Inactivation of the nucleotide excision repair prevents strand fragmentation after UV irradiation (100 J/m2), as revealed by alkaline sucrose gradient. Chromosomal DNA in this case was labeled chronically (the pre-labeling with 3H-thymidine protocol). The cells are: UvrA+, LA55; uvrA, LA44. Inset: the (A600=0.45) cultures of cells of the indicated phenotype were irradiated with the indicated dose of UV, and after 15' at 42°C the total DNA was isolated, run on an 0.7% alkaline agarose gel, that was transferred, and the membrane hybridized with the total chromosomal probe. C. Inactivation of the major abasic site endonuclease ExoIII by the xthA mutation prevents strand fragmentation after H2O2 treatment (0.5 mM for 13 minutes at 42°C) of chronically-labeled cultures. The “pre-labeling with 3H-thymidine protocol” was again used. Neutral sucrose gradient in this case, with samples boiled for 10 minutes and chilled on ice before loading. The strains are: ligA uvrA mutS, LA53; ligA uvrA mutS xthA, LA44. Inset: survival of the indicated strains (A600=0.4) after treatment with 1 mM H2O2 for 10 minutes at 30°C. D. Inactivation of mismatch removal prevents strand fragmentation after 2-aminopurine (2AP) treatment of the dam mutants. Cells of the indicated genotype were kept at 30°C throughout the assay. Once the cultures reached OD600= 0.2, they were labeled with 32P for 30 minutes, shaken in the presence of the indicated concentration of 2-AP overnight, the total DNA was prepared and ran in an alkaline agarose gel. Strand fragmentation was calculated as the percentage of DNA signal in the smear below the chromosomal DNA band relative to the total DNA signal (for example, see Fig. 5B and E). Since the background in untreated cells varied widely in this set of experiments, while the signal-to-background difference (including one negative value) was quite reproducible, we report the latter. The values are means of 4-5 independent measurements ± SEM. The strains are: ligA uvrA dam (LA73) and ligA uvrA mutsS dam (LA74). E. Sensitivity of the ligA uvrA dam strain to 2AP and its relief by the mutS defect. Cultures were shaken at 28°C throughout. When they reached OD600 = 0.4, 2AP was added to the indicated concentrations, and shaking continued for five more hours, at which time the titer of the colony-forming units was determined. The strains are: ligA uvrA dam (LA73) and ligA uvrA mutsS dam (LA74). F. Replication intermediates in the LigA+ mutant devoid of the major DNA excision activities are HMW independent of the denaturation conditions. Both neutral and alkaline sucrose gradients are shown in the same panel. The strain is LA115. G. Replication intermediates in the ligA251 mutant devoid of the major DNA excision activities are LMW independently of the denaturation conditions. Both neutral and alkaline sucrose gradients are shown in the same panel. The strain is LA114.